The Gena Dashboard is a Streamlit application designed for digital twin analysis and visualization. It provides an interactive interface for processing, analyzing, and interpreting digital twins through various bioinformatics workflows.
The aim is to simplify the use of the Gena Brick by providing an application that makes running the pipeline and retrieving results easier. Dependencies between scenarios are also maintained, allowing you to navigate more easily.
Multi-Step Analysis Pipeline
The dashboard implements a structured analysis workflow with the following steps:
- Network: Create a network and enable users to modify it through various tasks: Network, NetworkImporter, GapFiller, IsolateFinder, LoadBiggModels, NetworkMergem, NetworkMerger, OrphanRemover, ReactionAdder, ReactionRemover, TransporterAdder,
How to start?
Open a new scenario and add the Generate Gena Dashboard app task. Then, configure the parameters: choose whether each scenario must be associated with a folder in your space, and you can add the credentials of a larger lab. This could be useful for the final step, '16S'. Finally, run the task and open the Streamlit resource.
This dashboard enables you to perform new analyses and navigate through previous results.
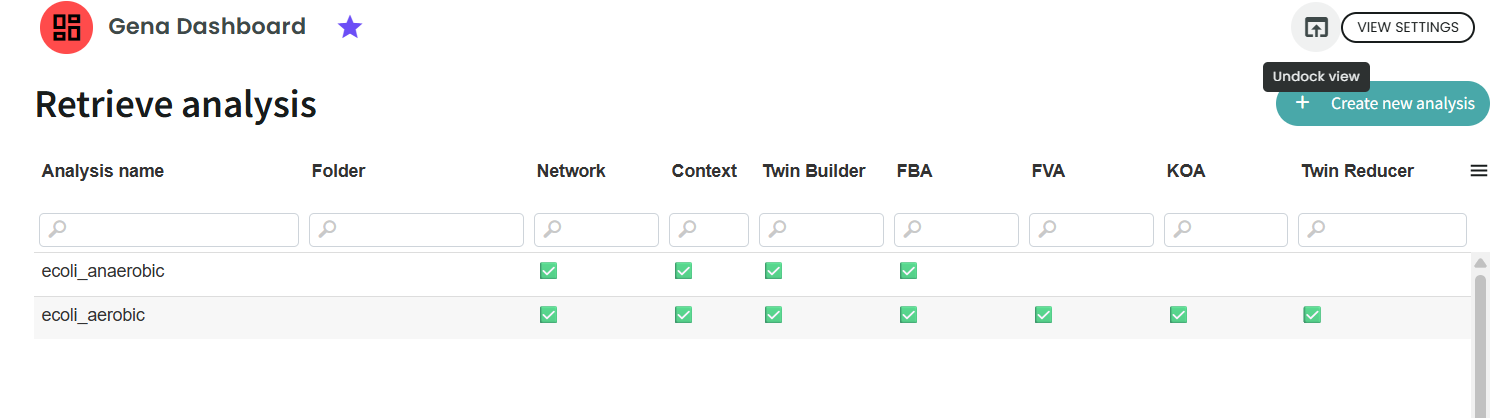
On the first page, you will see an overview of what has already been run and the status of each step.
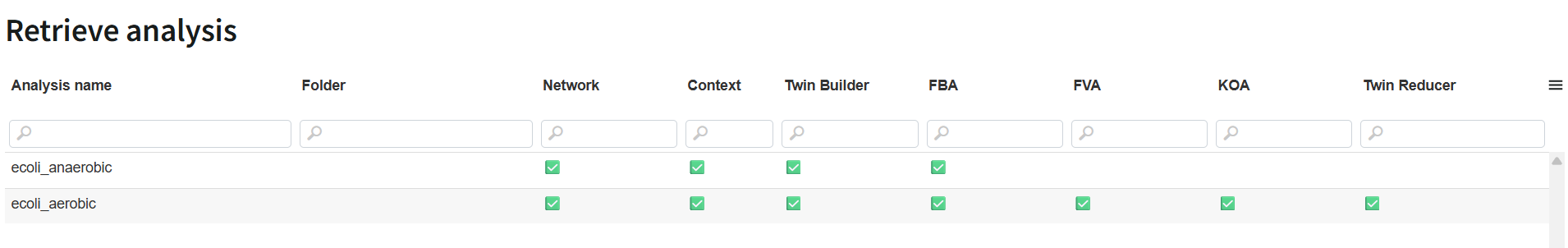
You can also click on the button "Create new analysis" to run a new analysis.
Once you have clicked on an analysis, you will see the details of each step and have the option to launch some analyses.
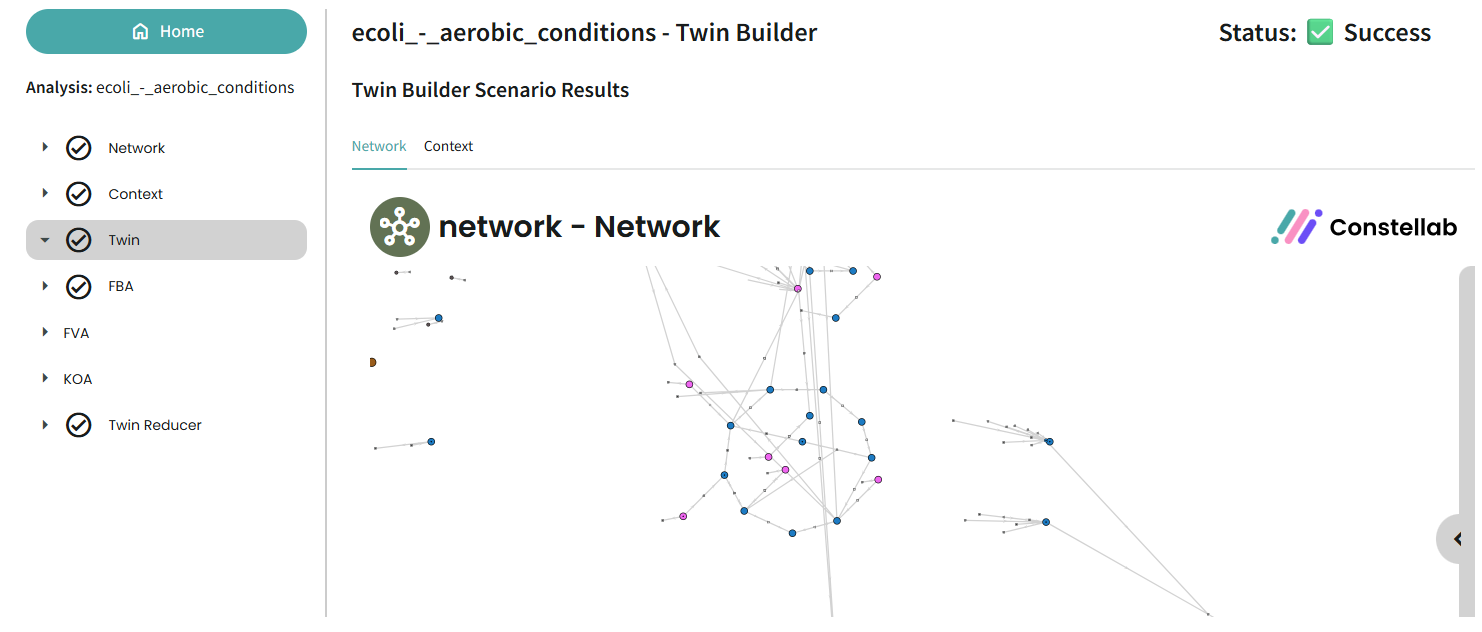
Network
Metabolic networks are highly flexible and can be modified through many different tasks — it’s a real playground for exploration. Thanks to interactive tools, you can immediately visualize changes in the network view, making it easier to refine and validate your model step by step.
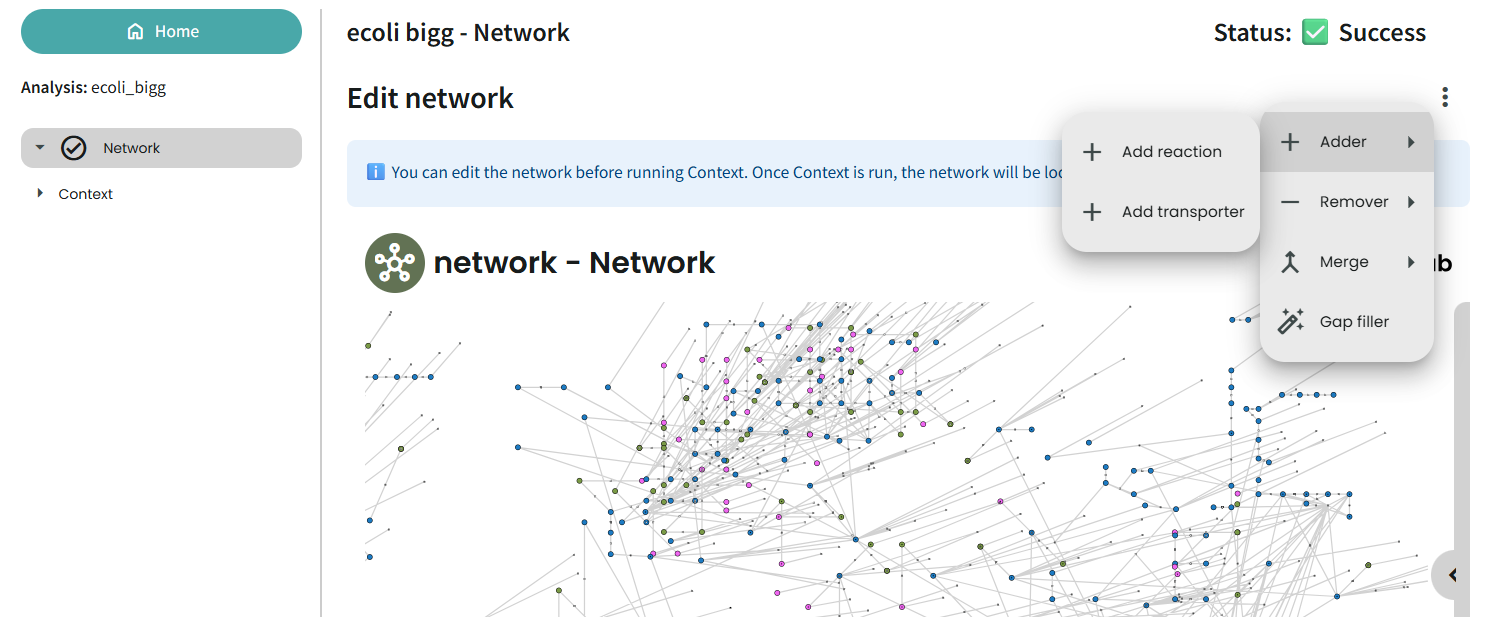
Best Practices
Network Preparation
- Validate networks before analysis: ensure the model structure follows the required format and passes basic integrity checks.
- Remove duplicate reactions and metabolites: redundant entries can lead to incorrect flux distributions and inconsistencies.
- Check for mass and charge balance: all reactions should be stoichiometrically balanced to avoid generating thermodynamically impossible results.
- Verify connectivity: confirm that metabolites are connected to the network and that no isolated sub-networks remain.
Context Definition
- Use realistic medium compositions: define uptake and secretion rates based on the actual experimental setup.
- Set appropriate flux bounds: avoid arbitrary limits; instead, use physiologically meaningful ranges.
- Validate experimental constraints: integrate experimental data carefully, ensuring it is consistent with the network.
Performance Optimization
- Use model reduction for large networks: simplify networks while preserving essential behavior to improve computational efficiency.
- Limit FVA to essential reactions: running FVA across the entire model can take time; focus on reactions of interest.
FAQ
Q: How should I begin?
Click 'Create new analysis' to launch your first scenario. This step involves taking a network or import one from BiGG Models and running the Metadata task. Once this step has been completed successfully, you can click on the first analysis in the table to view the metadata results, modify the table and run the other steps!
Q: How can I view my entire analysis?
Simply click on the desired analysis in the table on the home page and you will be redirected to the analysis details, where you can see all the steps.
Q: One of my scenarios is showing an error. What should I do?
Click on "view scenario in your dashboard", you will be redirected to the scenario in your lab. You will then see details of the error. Try to resolve the issue by adjusting the parameters or input. Please be careful not to delete the output.
If you don't know how to modify the scenario, note that you still can delete this one and create a new one though the dashboard.

