Introduction
The use case presents main steps to create and prepare a metabolic network of an organism without data or with custom core genome information. It also presents how to visualise and navigate though the network and extract meaningful information on the model.
Final networks can be used to simulate and predict the metabolism of the organism in different context (custom cell culture, Gene KO).
Metabolic network reconstruction
Building of a metabolic model using genome data
If you have genomic data from your organism to reconstruct, you can use Diamond and EC number tasks. See the documentation here.
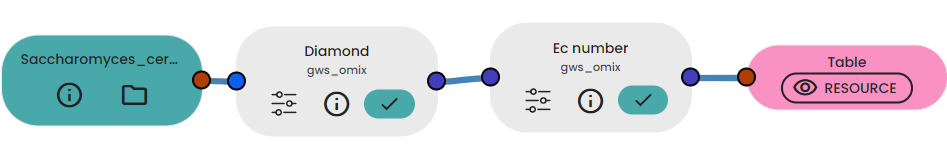
Using the table of EC numbers extracted from the first part, we can use it to reconstruct an entire metabolic model network from scratch!
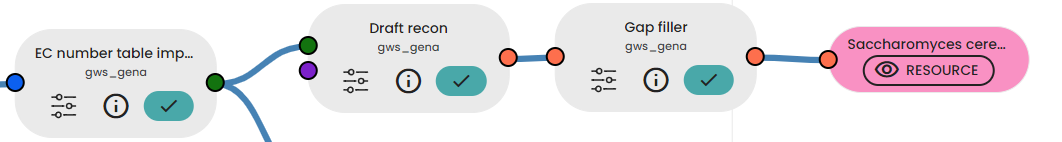
You need to link the table of EC numbers to the 'Draft recon' task, which aims to reconstruct a metabolic network. If you have a biomass reaction table that describes the composition of the biomass of a cell (or organism), you can also add it to the second input. This additional information will then be added to your network.
You can then connect this draft metabolic network to the Task "Gap Filler". This task iteratively fills gaps related to dead-end compounds using the Biota database. A gap is detected when a steady compound is a dead-end compound.
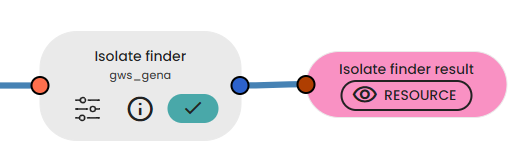
As an additional step, the Isolate Finder task can be used to find isolate reactions/compounds that are not related to cell growth. Take a close look at these reactions and compounds as you will certainly need to remove them from your model!
After these automatic tasks and manual checks, you should now have a robust metabolic model that can be used in Constellab for further analysis.
Building of a metabolic model without data
If you don't have genomic data for your organism, don't worry, we'll look at other methods for reconstructing a genome-scale metabolic model. Indeed, you can set the parameter "tax_id" and the parameter "tax_search_method" to define the search method.
In the output, you will have a draft of a metabolic model and you can apply the same Tasks as before.
Visualise the network
In Constellab you can have easy access to the information contained in your network.
Visualise the network
In Constellab, you will obtain this view:
You can click on the right panel to see many options: decide which pathway to display, get information about a reaction/metabolite...
Number of compounds and reactions
With the JSON view, you can easily see the number of metabolites, reactions, genes ...
Mass and charge balance of each reactions
In the previous view, if you unfold the panel a few times, you can see precise information about a metabolite/reaction, such as the charge balance, the mass, bounds, compartment ...
Conclusion
With the help of Constellab, we can use this complete pipeline to build a metabolic network simply from genomic data or taxonomy ID and make it robust by gap filling, isolate finder and manually check, if necessary. The different views in Constellab make it easy to visualise and use your model.

