Introduction
In the dawn of a genomic revolution, where the blueprints of all known species are meticulously unraveling through comprehensive sequencing efforts, the demand for advanced tools becomes paramount. As we traverse this era of data-driven biology, one standout protagonist emerges – DIAMOND, a cutting-edge tool poised to redefine the landscape of genomic exploration (click here).
Picture a world where the intricate language of DNA, the code of life itself, is deciphered for every organism on Earth. In this vast repository of genetic information, comparative analyses hold the key to unlocking mysteries and revealing the intricate connections that bind all living beings. Enter DIAMOND, an evolutionary leap in bioinformatics, presenting an enhanced version that transcends the boundaries of its predecessors while matching the sensitivity of the gold standard BLASTP.
This tool uses Blastp and UniProtKB/Swiss-Prot database.
The obtained results pave the way for a crucial next step – the determination of Enzyme Commission (EC) numbers for the reconstruction of digital twins.
Prerequesites
Steps to follow
- Upload your FASTA file in your databox.
- Link it to the task "Diamond" available in the brick gws_omix.
- Link the output of Dimond task to "Ec number" task available in the same brick.
- Fill parameters : you must indicate the type of input sequence (nuc or prot) , the coverage and the e-value.
- Run your experiment.
- Get your Blast result and EC number in 2 different tables representing the outcome of distinct tasks.
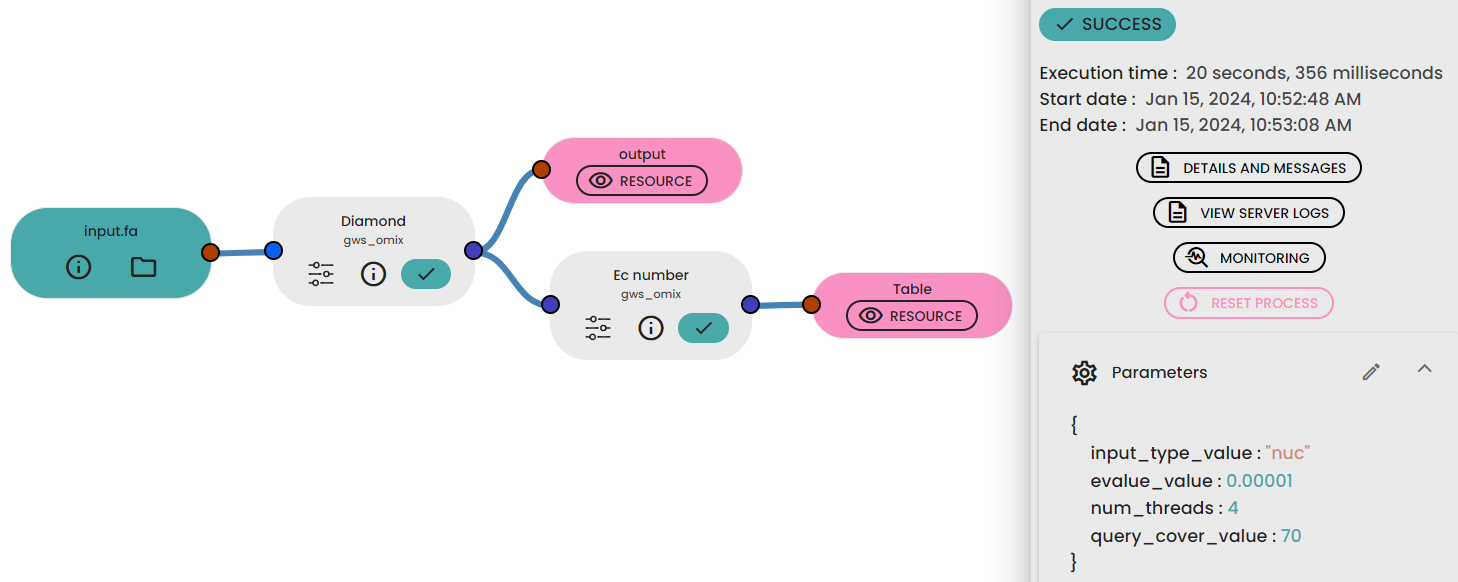
Results
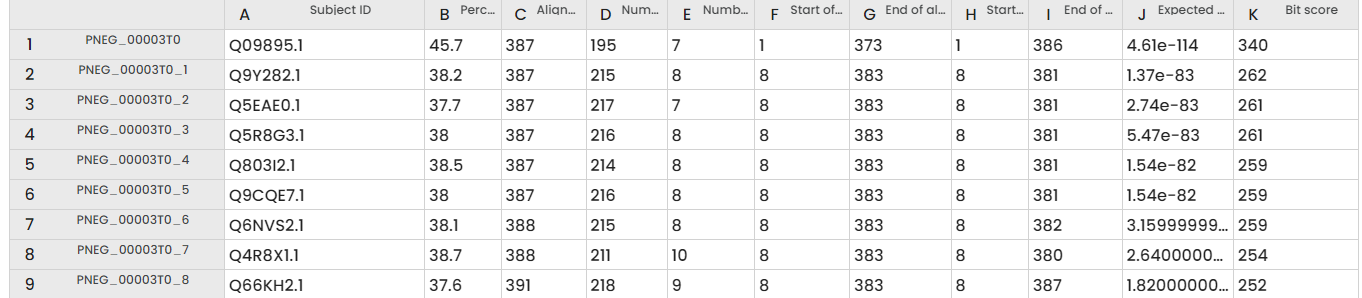
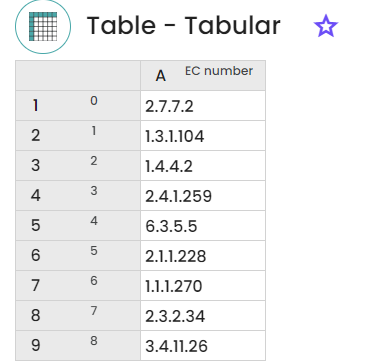
The experiment yields two resources: firstly, a table displaying the subject ID, bit score, e-value (as depicted in the figure below); and secondly, a table representing the EC numbers.

Comments (0)
Write a comment