Introduction
Once armed with a genome-scale metabolic model, the pivotal next step lies in unleashing the power of Flux Balance Analysis (FBA). By analyzing and optimizing the flow of metabolites through this metabolic landscape, FBA unveils insights into cellular behaviors, helping researchers understand how an organism allocates resources, maximizes growth, and adapts to diverse environmental conditions. FBA can be performed on Constellab.
Once you have run the FBA on the two conditions you want to compare, it can be difficult to know what to do next. A first step might be to compare the simulated fluxes and plot a graph. Let's do it!
Prerequesites
You will need two flux tables from an FBA task. If you have contextualised your model to perform the PCA, you will need to provide a dataframe containing the modified reactions.
Steps to follow
- Upload your two flux tables and a dataframe containing the reactions modified.
- Link it to the task available in the brick.
- Fill parameters : you must specify the name of the conditions being compared, the column containing the modified reactions and finally the threshold. In the advanced configuration you can specify if you want a logarithmic axis on x and/or y.
- Run your experiment.
- Get your coloured plot and the dataframe created.
Results
This tutorial was created using the E.coli model from BiGG. We previously conducted two FBAs in two different contexts. The 2 resulting flux tables are attached to this story along with the list of modified reactions.
Once you have run the task, you will get these two outputs:
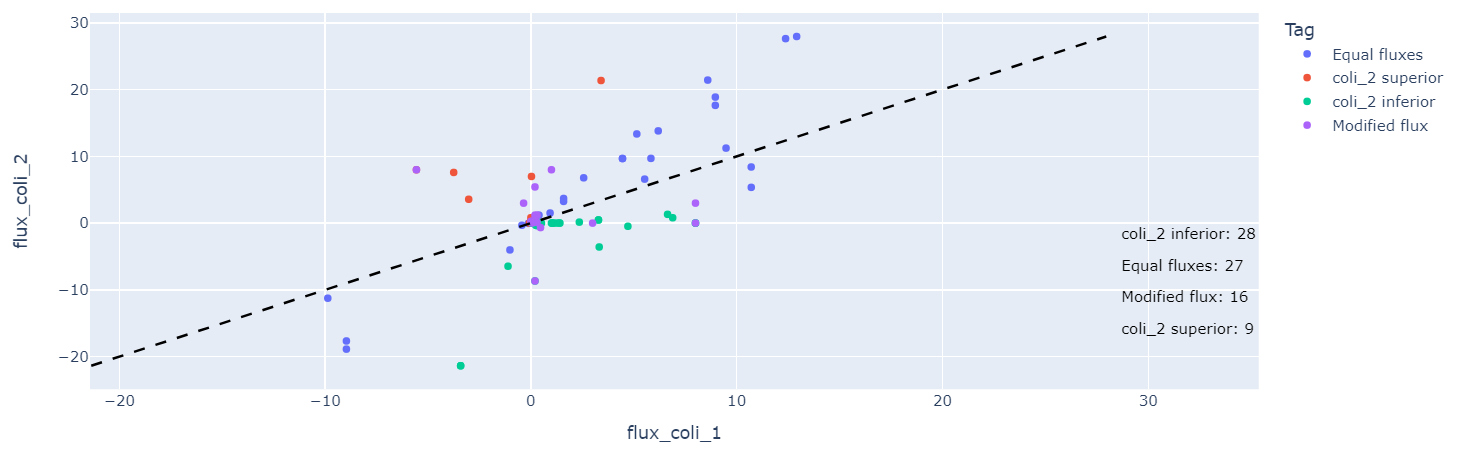
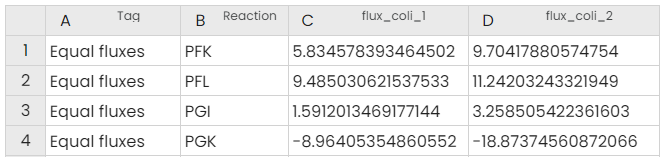
The output table can be used to go further. In fact, you can do further analysis, such as preserving the reactions marked as superior and extracting the genes involved in these reactions. These genes can be studied to know if certain pathways are more affected. You can do gene enrichment thanks to websites: DAVID, ShinyGO... Either way you can also continue on Constellab by using the Task KEGG Visualisation (see the story here).

Comments (0)
Write a comment