Introduction
Ever since the beginning of the 21st century, we have witnessed remarkable technological advancements in biology, particularly with the emergence of high-throughput ‘omics’ techniques such as microarrays and next-generation sequencing (NGS). These approaches revolutionized genomics by enabling the simultaneous measurement of thousands of genes in a single experiment. As a result, researchers can now rapidly identify genes whose expression is associated with particular phenotypes, treatments, or conditions.
However, identifying differentially expressed genes (DEGs) is only the first step. The true challenge lies in interpreting these long gene lists in a biologically meaningful way. A list of hundreds or thousands of genes provides little insight on its own; instead, scientists need to understand which biological processes, pathways, or molecular mechanisms are perturbed. To address this challenge, a variety of computational approaches have been developed to translate raw gene-level information into interpretable functional categories. This family of approaches is collectively known as Functional Enrichment Analysis (FEA) or Gene Set Analysis (GSA).
The central aim of FEA is to determine whether certain biological annotations such as pathways, molecular functions, or cellular components are statistically overrepresented in a gene set of interest relative to an appropriate reference background. By linking DEGs to known biological knowledge bases (e.g., Gene Ontology (GO), KEGG pathways, or Reactome), FEA allows researchers to move from raw lists of genes to higher-level interpretations about the underlying biological mechanisms in their study system.
ORA = I have a shortlist => is this pathway overrepresented in it?
GSEA = Across the whole ranked list => do the genes from a pathway cluster toward the top or the bottom?
Methods
1. Overrepresentation Analysis (ORA)
1.1 What's ORA?
ORA is one of the most widely used and conceptually simple methods for functional enrichment. Here, the input is a discrete list of genes typically DEGs identified after applying thresholds on adjusted p-values and fold-changes.
The question ORA asks is:
Do certain pathways or functional categories contain more genes from my list than would be expected by chance?
This is statistically tested using the hypergeometric test, which assesses whether the overlap between the gene list and a given pathway is greater than random expectation.
One important consideration in ORA is the definition of the background universe. By default (auto mode), the background is set to the entire reference genome (e.g., ~20,000 genes in humans). Alternatively, in experiment mode, the background can be defined as all genes tested in the experiment (e.g., ~12,000 measured transcripts in RNA-seq). The choice of background influences the results, especially in tissue or platform specific datasets.
1.2 Tools
While several popular R packages exist for functional enrichment analysis, such as topGO and clusterProfiler, the g:Profiler ecosystem (including the R client gprofiler2) offers distinct advantages. Unlike tools that require local downloads of multiple annotation databases, g:Profiler provides access to a broad range of high-quality, regularly updated annotation sources through a single query. Its in-house database integrates only well-established and reliable resources, including Gene Ontology (GO), KEGG, Reactome, WikiPathways, miRTarBase, TRANSFAC, the Human Protein Atlas, protein complexes from CORUM, and the Human Phenotype Ontology.
Another important feature is its automatic identifier mapping: g:Profiler seamlessly handles input lists containing mixed types of gene identifiers (e.g., Ensembl IDs, gene symbols, Entrez IDs), removing a common source of preprocessing errors. This flexibility, combined with continuous updates and support for over 600 species and strains, makes g:Profiler an ELIXIR Recommended Interoperability Resource in life sciences. Overall, g:Profiler provides a user-friendly, reproducible, and comprehensive framework for conducting functional enrichment analysis without the technical burden of managing multiple annotation sources locally.
1.3 Steps
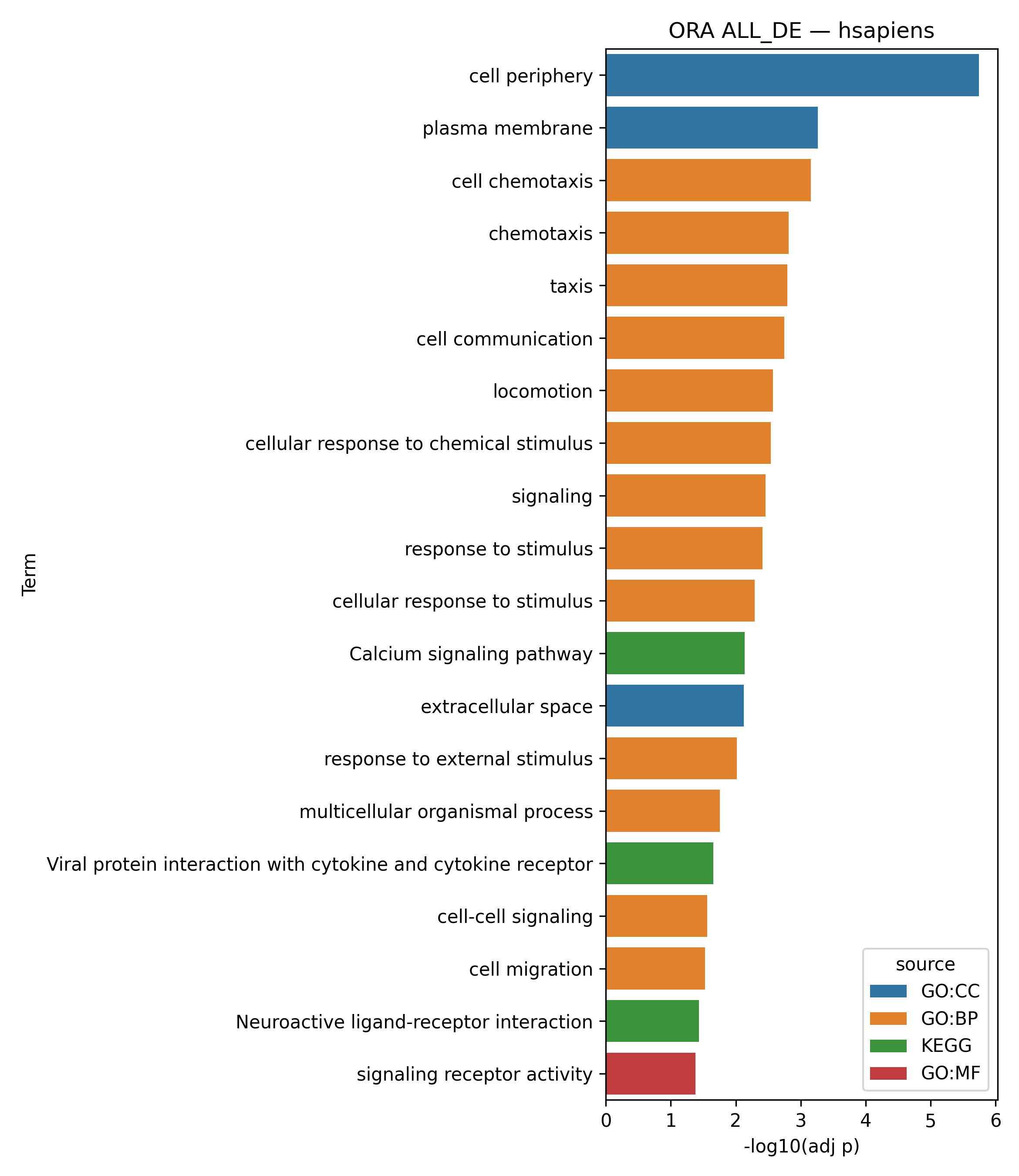
This task (task: OmiX – Functional enrichment analysis based on ORA) performs functional enrichment analysis on the list of differentially expressed genes (DEGs). It conducts Over-Representation Analysis (ORA) using a hypergeometric/Fisher’s exact test, followed by multiple-testing correction (by default via g:SCS – Set Counts and Sizes).
In this workflow, the background universe is set to auto (the whole genome as defined by g:Profiler) rather than experiment (all genes quantified in the dataset). This choice ensures consistency with the g:Profiler web interface, where only terms passing the corrected significance threshold (adjusted p-value ≤ α) are retained.
Parameters:
Outputs