Introduction
Biomath, the quiet engine behind great experiments
Every successful wet-lab workflow is powered by a handful of invisible, high-stakes decisions: how many molecules am I adding? What’s the true concentration after dilution? Is my insert:vector ratio correct? How does an OD reading translate into actual nucleic acid mass? These are not trivial details they’re the difference between a crisp gel and a “what went wrong?” day.
Biomath calculators turn that uncertainty into clarity. They bridge instruments and intuition, translating absorbance into µg/ml, mass into pmol, stock into working solutions, and DNA length into coding capacity. With a few keystrokes, you can standardize mixtures, make ligations reproducible, size constructs realistically, and communicate results in the units that matter for biology: molecules, molarity, mass, and temperature.
Why it matters:
- Reproducibility: consistent molar inputs give consistent outcomes.
- Efficiency: fewer back-of-the-envelope errors, fewer failed runs.
- Interoperability: results you can share across teams, instruments, and protocols.
- Design power: plan constructs and reactions with confidence, from PCR to protein work.
In short: biomath is not busywork it’s the foundation of good experimental design. The tools in this suite are fast, transparent, and grounded in well-vetted formulas so you can spend less time guessing and more time discovering.
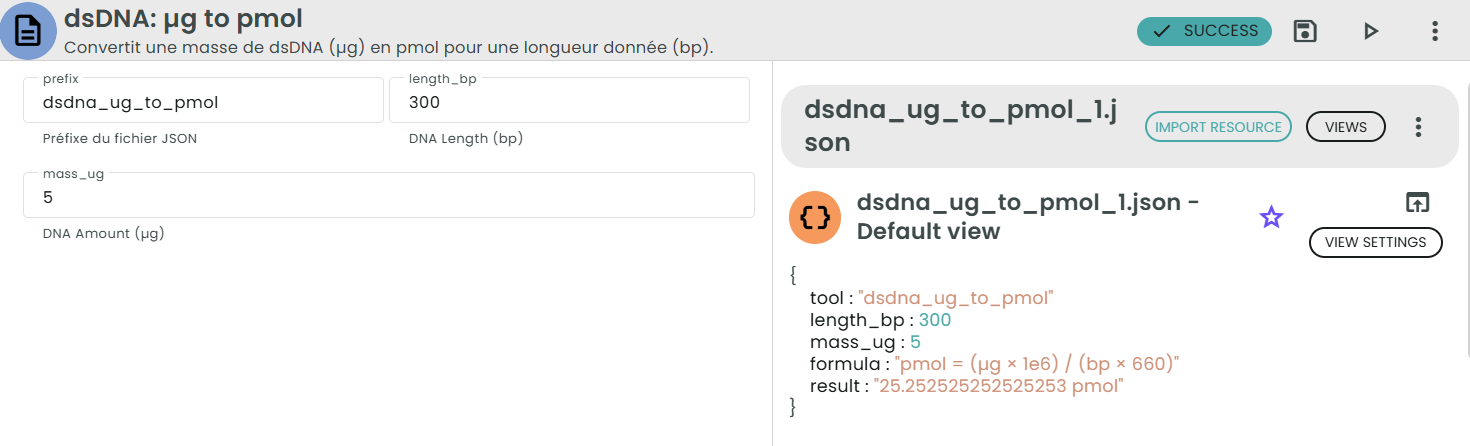
✨dsDNA: µg → pmol✨
What: Convert mass of double-stranded DNA (µg) to amount in picomoles (pmol).
Why: You need pmol to set molar ratios for ligations, assemblies, and equimolar mixes.
Formula:
pmol = (µg × 1e6) / (bp × 660)
Where:
• 1e6 is the conversion factor from micrograms (µg) to picograms (pg).
• 660 pg/pmol is the average molecular weight of one base pair of double-stranded DNA.
• bp is the length of the DNA fragment in base pairs.
Example:
For 1 µg of a 3000 bp fragment:
pmol = (1 × 1e6) / (3000 × 660) = 0.505 pmol
Interpretation:
0.505 pmol corresponds to approximately 3.04 × 10^11 DNA molecules,
because 1 pmol = 6.022 × 10^11 molecules (Avogadro’s number).
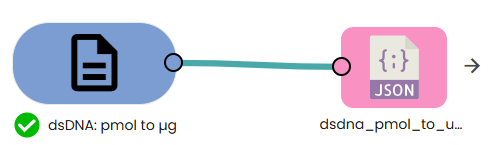
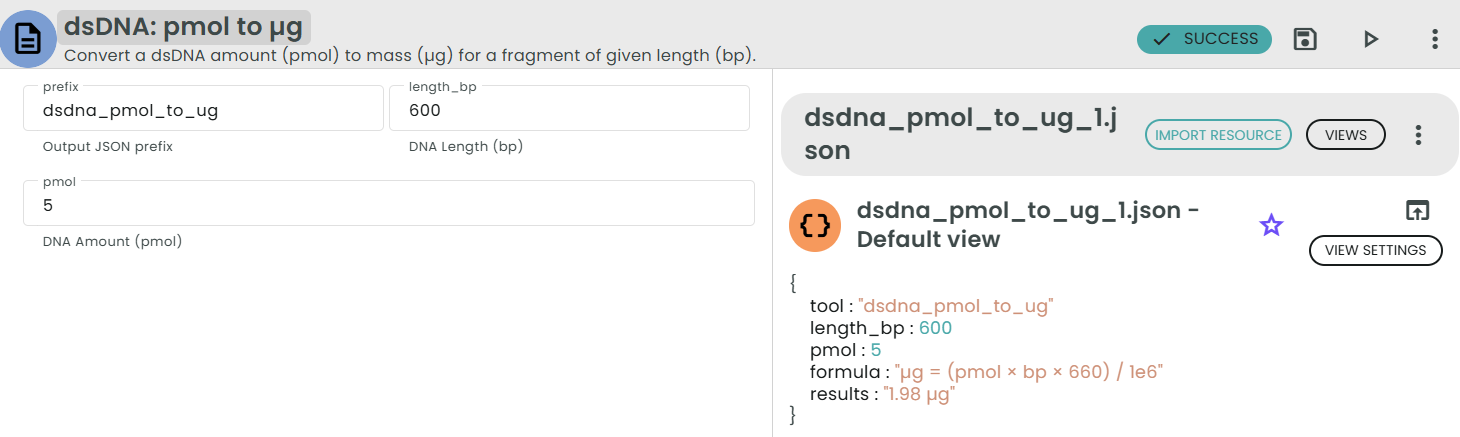
✨dsDNA: pmol → µg✨
What: Convert picomoles (pmol) of dsDNA to mass (µg).
Why: Useful for preparing a target mass from a desired number of molecules.
This module converts a molecular amount of double-stranded DNA (pmol) into a mass (µg) for a DNA fragment of a given length in base pairs (bp). In other words: Given N base pairs and a certain amount in picomoles, what is the corresponding mass in micrograms?
Formula:
µg = (pmol × bp × 660) / 1e6
Where:
• 660 pg/pmol is the average molecular weight of one base pair of dsDNA.
• 1e6 converts picograms (pg) to micrograms (µg).
• bp is the fragment length in base pairs.
Example:
For 1 pmol of a 3000 bp fragment:
µg = (1 × 3000 × 660) / 1e6 = 1.98 µg
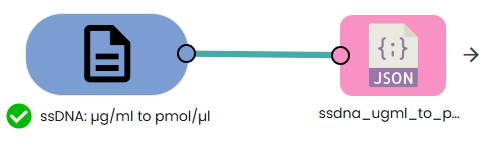
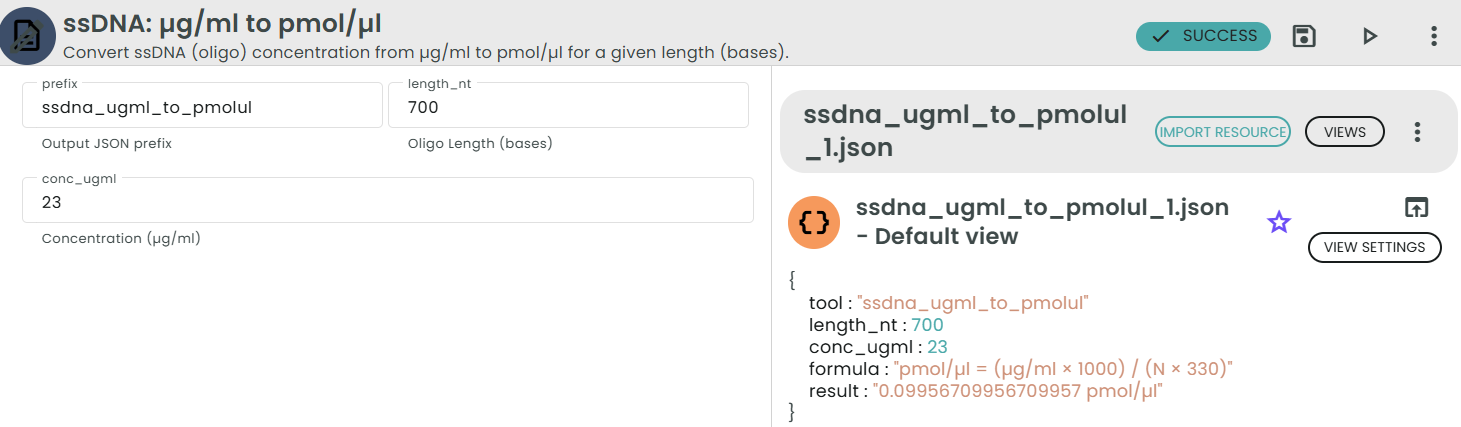
✨ssDNA: µg/ml → pmol/µl✨
What: Convert oligo concentration from µg/ml to pmol/µl.
Why: PCR oligo setup and dilutions are typically done in molar units.
This module converts a single-stranded DNA (oligo) concentration from micrograms per milliliter (µg/ml) to picomoles per microliter (pmol/µl), given the oligo length in nucleotides (bases).
Formula:
pmol/µl = (µg/ml × 1000) / (N × 330)
Where:
• N is the oligo length (number of nucleotides, bases).
• 330 pg/pmol is the average molecular weight of a single nucleotide for ssDNA.
• 1000 converts milliliters to microliters (1 ml = 1000 µl).
Derivation:
(µg/ml) × (ml / 1000 µl) × (10^6 pg / 1 µg) × (1 pmol / 330 pg) × (1 / N)
= (µg/ml) × 1000 / (330 × N) pmol/µl
Example:
For an oligo of 25 bases at 10 µg/ml:
pmol/µl = (10 × 1000) / (25 × 330) ≈ 1.212 pmol/µl
✨ssDNA: pmol/µl → µg/ml✨
What: Convert oligo concentration from pmol/µl to µg/ml.
Why: Some specs and instruments report mass concentration; convert back from molarity.
This module converts a single-stranded DNA (oligo) concentration from picomoles per microliter (pmol/µl) to micrograms per milliliter (µg/ml), given the oligo length in nucleotides (bases).
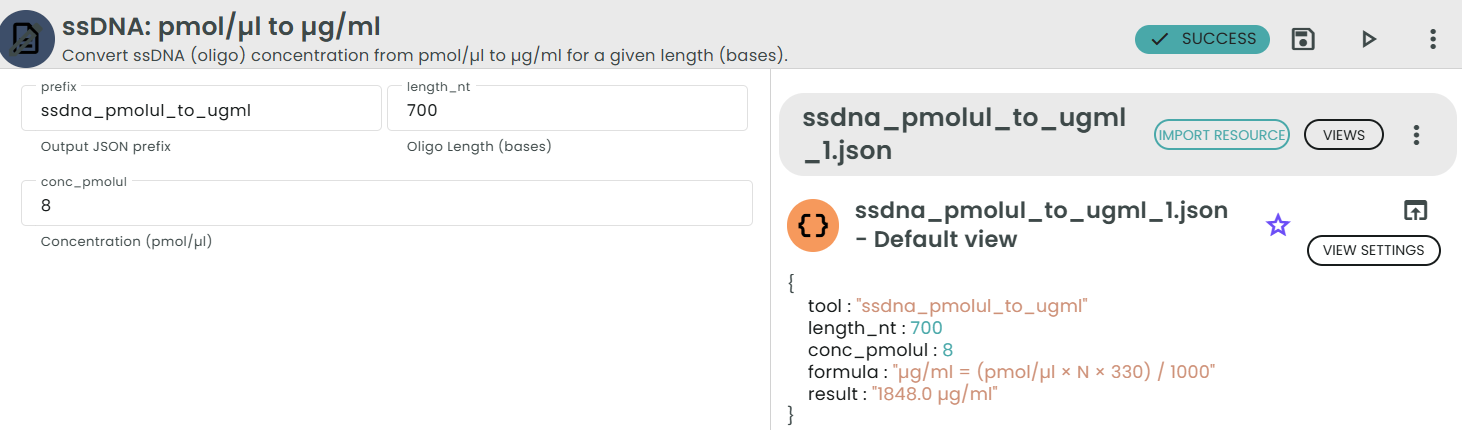
Formula:
µg/ml = (pmol/µl × 1000 µl/ml × 330 pg/pmol × N) × (1 µg / 1e6 pg)
= (pmol/µl × N × 330) / 1000
Where:
• N is the oligo length (number of nucleotides, bases).
• 330 pg/pmol is the average molecular weight of a single nucleotide for ssDNA.
• 1000 converts microliters to milliliters (1000 µl = 1 ml).
• 1e6 converts picograms to micrograms (1 µg = 10^6 pg).
Example:
For a 25-base oligo at 1.212 pmol/µl:
µg/ml = (1.212 × 25 × 330) / 1000 ≈ 10.0 µg/ml
✨Linear DNA: µg → pmol of Ends✨
What: Convert mass of linear dsDNA to pmol of DNA ends available.
Why: Ligation and end-labeling stoichiometry depend on the number of ends.
This tool converts the mass of a linear double-stranded DNA (dsDNA) fragment (in micrograms, µg) to the picomoles of DNA ends available, given the fragment length in kilobases (kb).
Rationale:
• Average mass per base pair (bp) of dsDNA ≈ 660 pg/pmol.
• Convert length from kb to bp: bp = kb × 1000.
• Picomoles of DNA molecules:
pmol_molecules = (µg × 1e6 pg/µg) / (bp × 660 pg/pmol).
• A linear dsDNA molecule has two ends:
pmol_ends = pmol_molecules × 2.
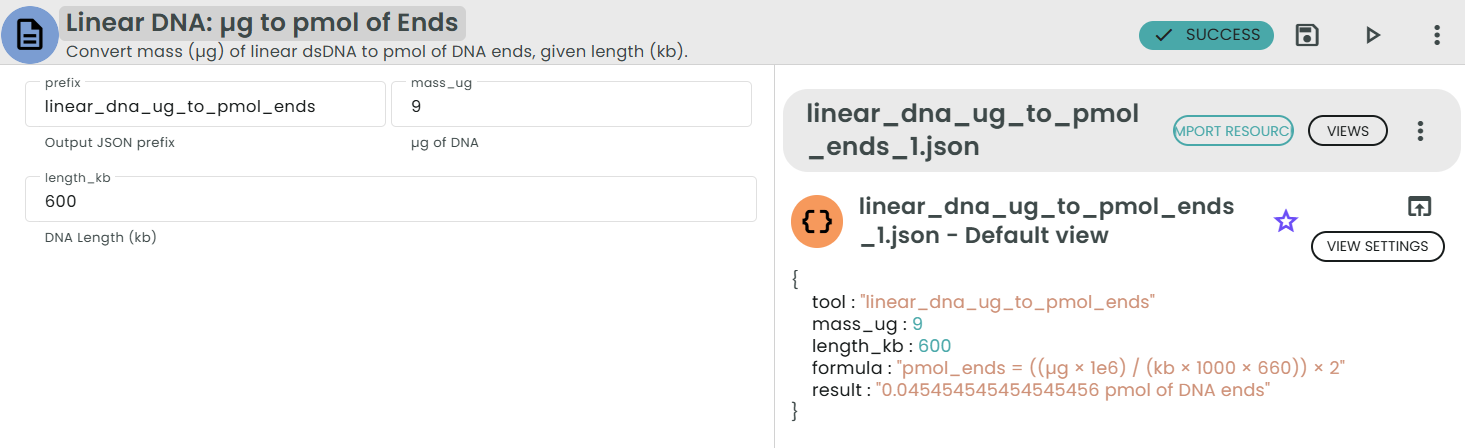
Compact formula:
pmol_ends = (µg × 1e6) / (kb × 1000 × 660) × 2
= (µg × 2 × 10^6) / (kb × 660 × 1000)
Example:
For 0.5 µg of a 4 kb linear dsDNA fragment:
bp = 4 × 1000 = 4000
pmol_molecules = (0.5 × 1e6) / (4000 × 660) ≈ 0.189 pmol
pmol_ends = 0.189 × 2 ≈ 0.378 pmol of DNA ends
✨Ligations: Molar Ratio of Insert:Vector✨
What: Compute ng of insert needed for a given insert:vector molar ratio.
Why: Achieving optimal ligation efficiency depends on proper molar ratios.
This calculator determines how many nanograms (ng) of an insert DNA fragment are required to achieve a specific molar ratio with a given vector DNA fragment.
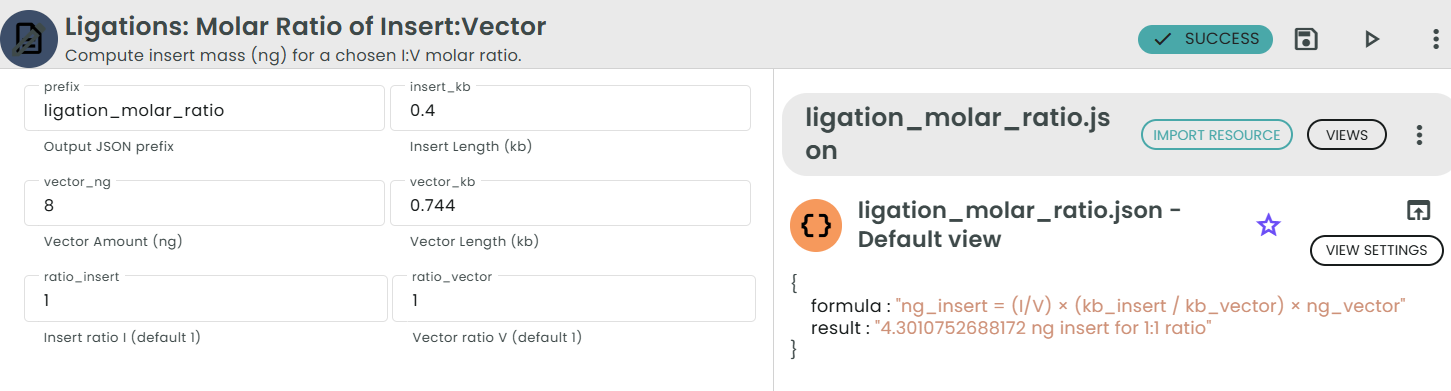
Formula:
ng_insert = (I / V) × (kb_insert / kb_vector) × ng_vector
Where:
• I is the desired molar ratio of insert (e.g., 1 for 1:1, 3 for 3:1).
• V is the molar ratio of vector (usually 1).
• kb_insert is the insert length in kilobases (kb).
• kb_vector is the vector length in kilobases (kb).
• ng_vector is the amount of vector DNA used in nanograms (ng).
Example:
For a 0.4 kb insert, a 0.744 kb vector, 8 ng of vector DNA, and a 1:1 ratio:
ng_insert = (1 / 1) × (0.4 / 0.744) × 8 ≈ 4.3 ng
✨Nucleic Acid: OD260 → µg/ml✨
What: Convert A260 (spectrophotometer) to nucleic acid concentration in µg/ml.
Why: Rapid quantification from absorbance without standard curves.
This calculator converts a nucleic acid absorbance at 260 nm (OD260) into concentration in micrograms per milliliter (µg/ml).
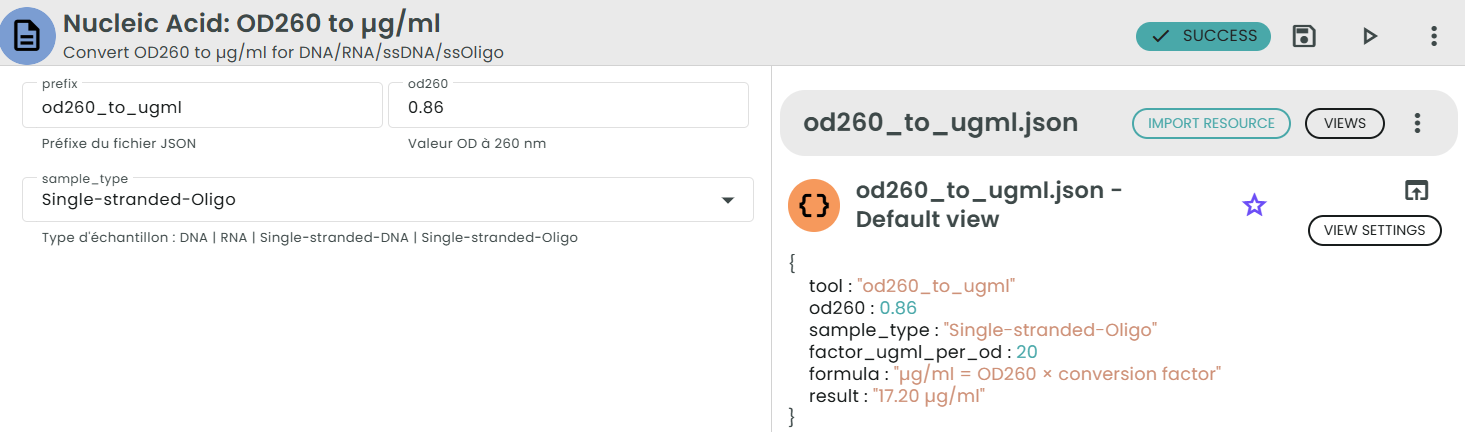
Formula:
µg/ml = OD260 × conversion factor
Conversion factors:
• DNA (dsDNA): 50 µg/ml per OD
• RNA (ssRNA): 40 µg/ml per OD
• Single-stranded DNA: 35 µg/ml per OD
• Single-stranded Oligo: 20 µg/ml per OD
Example:
For OD260 = 0.86 measured on a single-stranded oligo:
µg/ml = 0.86 × 20 = 17.2 µg/ml
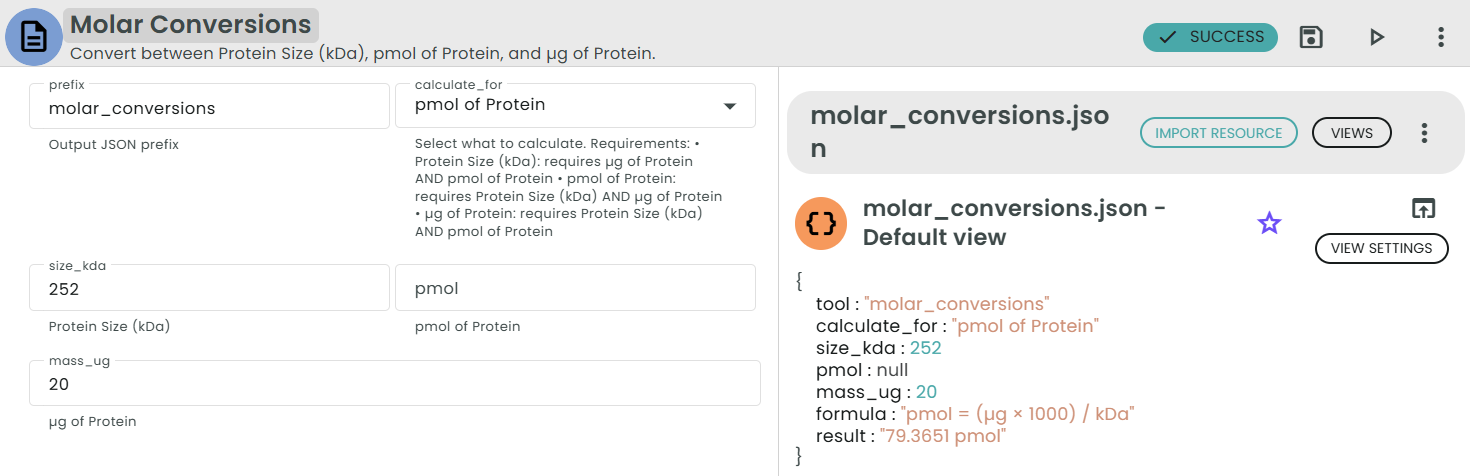
✨Molar Conversions (Protein)✨
What: Convert between protein size (kDa), pmol, and µg.
Why: Prepare equimolar protein mixes or normalize to molar input.
Description:
This module converts between:
• Protein Size (kDa)
• pmol of Protein
• µg of Protein
• µg protein = protein size (kDa) × pmol protein × (10^9 µg / kg) × (kg / 10^12 pmol)
• pmol protein = (µg protein / protein size (kDa)) × (10^12 pmol / mol) × (mol / 10^9 µg)
• protein size = (µg protein / pmol protein) × (10^12 pmol / mol) × (mol / 10^9 µg)
Note:
A protein that is 1 kDa has a molecular weight of 1 kilogram per mole.
Simplified formulas used by this tool:
• µg = (kDa × pmol) / 1000
• pmol = (µg × 1000) / kDa
• kDa = (µg × 1000) / pmol
Derivations (unit logic):
1) µg of Protein
MW in µg/mol = kDa × (1 kg/mol) × (10^9 µg / 1 kg) = kDa × 10^9 µg/mol
moles = pmol × (10^-12 mol / 1 pmol)
µg = (kDa × 10^9) × (pmol × 10^-12) = kDa × pmol × 10^-3 = (kDa × pmol) / 1000
2) pmol of Protein
pmol = (µg / (MW in µg/mol)) × 10^12
= (µg / (kDa × 10^9)) × 10^12
= (µg × 10^3) / kDa
→ pmol = (µg × 1000) / kDa
3) Protein Size (kDa)
kDa = MW / (10^9 µg/mol), and MW = (µg / pmol) × 10^12
kDa = [ (µg / pmol) × 10^12 ] / 10^9 = (µg × 10^3) / pmol
→ kDa = (µg × 1000) / pmol
How to use:
• If you choose: Protein Size (kDa)
- Fill in: µg of Protein, pmol of Protein
- Formula: kDa = (µg × 1000) / pmol
• If you choose: µg of Protein
- Fill in: Protein Size (kDa), pmol of Protein
- Formula: µg = (kDa × pmol) / 1000
• If you choose: pmol of Protein
- Fill in: Protein Size (kDa), µg of Protein
- Formula: pmol = (µg × 1000) / kDa
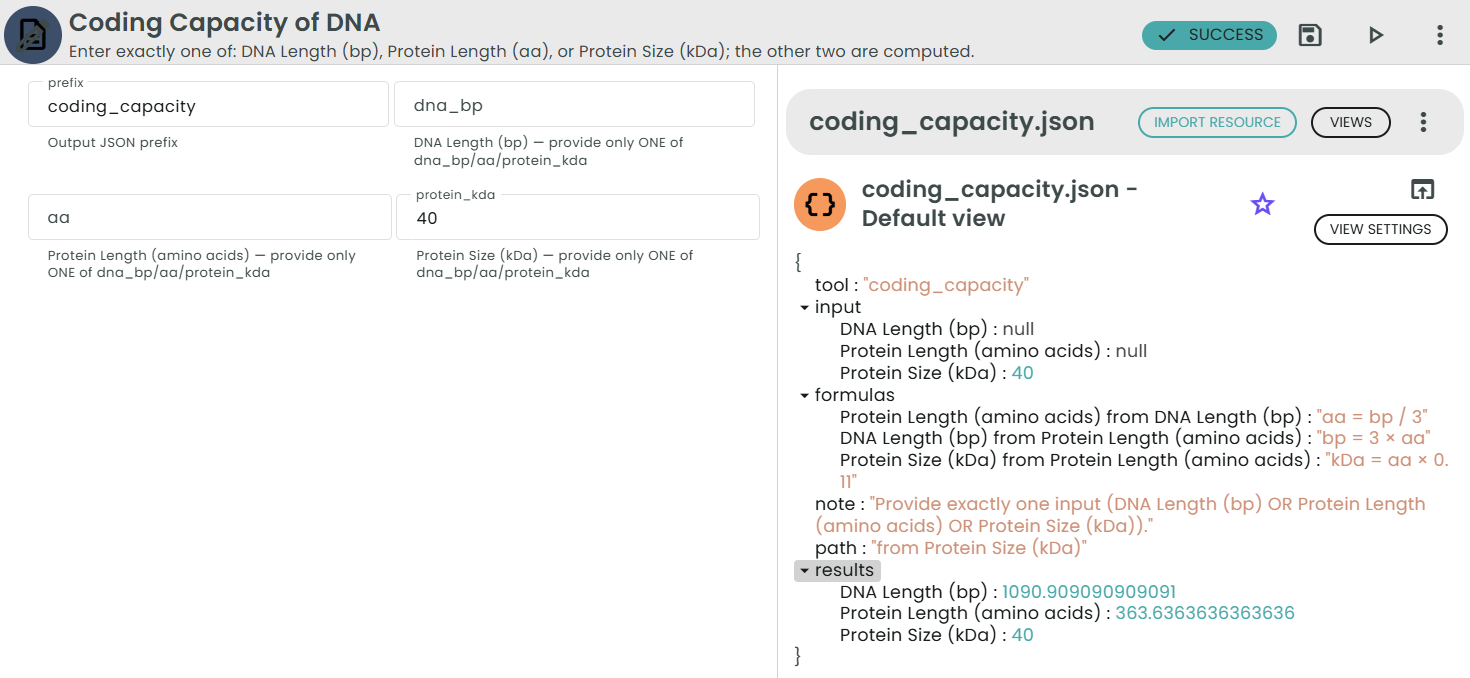
✨Coding Capacity of DNA✨
What: Convert between DNA size (bp), protein length (aa), and approximate protein size (kDa).
Why: Quick sense-check for gene sizes, construct design, or expected protein MW.
Enter exactly ONE parameter:
• DNA Length (bp), or
• Protein Length (amino acids), or
• Protein Size (kDa)
The tool computes the other two using:
• amino acids = DNA (bp) / 3
• DNA (bp) = 3 × amino acids
• protein size (kDa) = amino acids × 0.11
Notes:
• 0.11 kDa is the average molecular weight per amino acid (≈110 Da per residue).
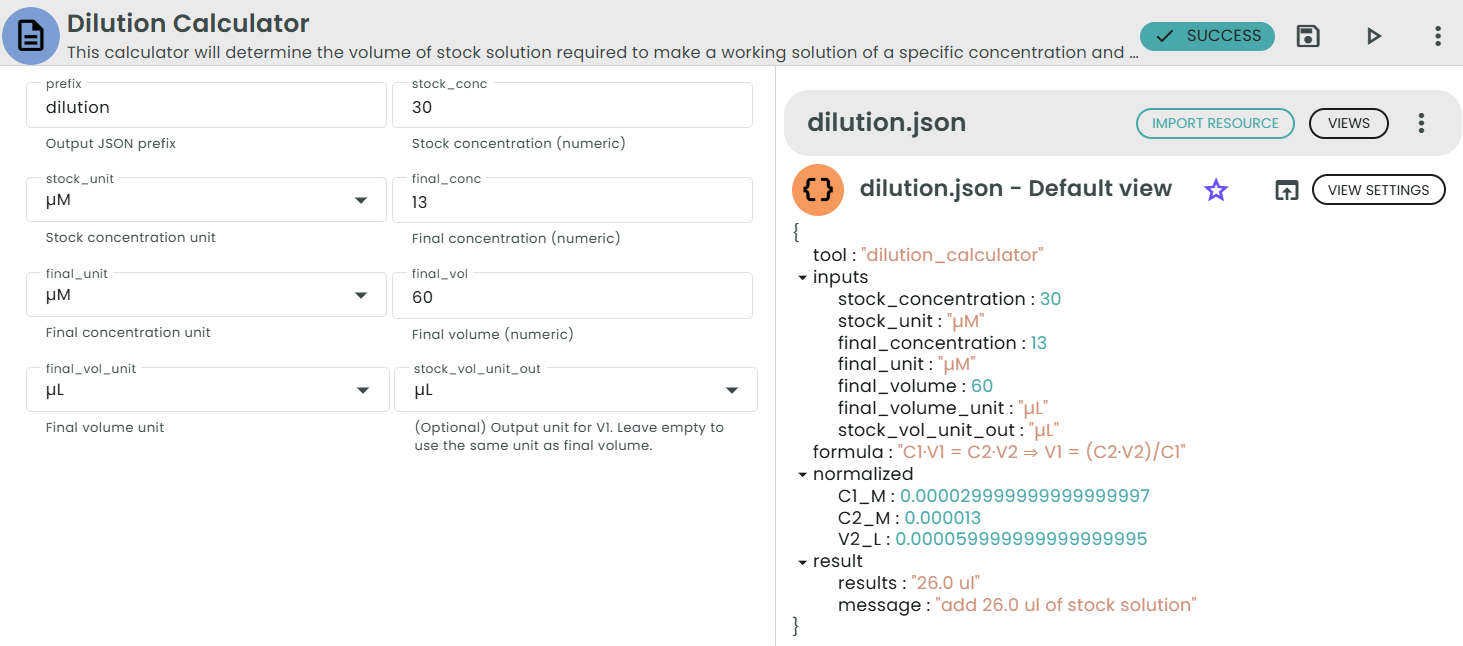
✨Dilution Calculator✨
What: Volume of stock needed to prepare a target concentration and volume.
Why: Routine lab dilutions rely on this identity.
Enter:
• C1 = Stock concentration
• C2 = Final (target) concentration
• V2 = Final (target) volume
Formula:
• C1 · V1 = C2 · V2
• V1 = (C2 · V2) / C1
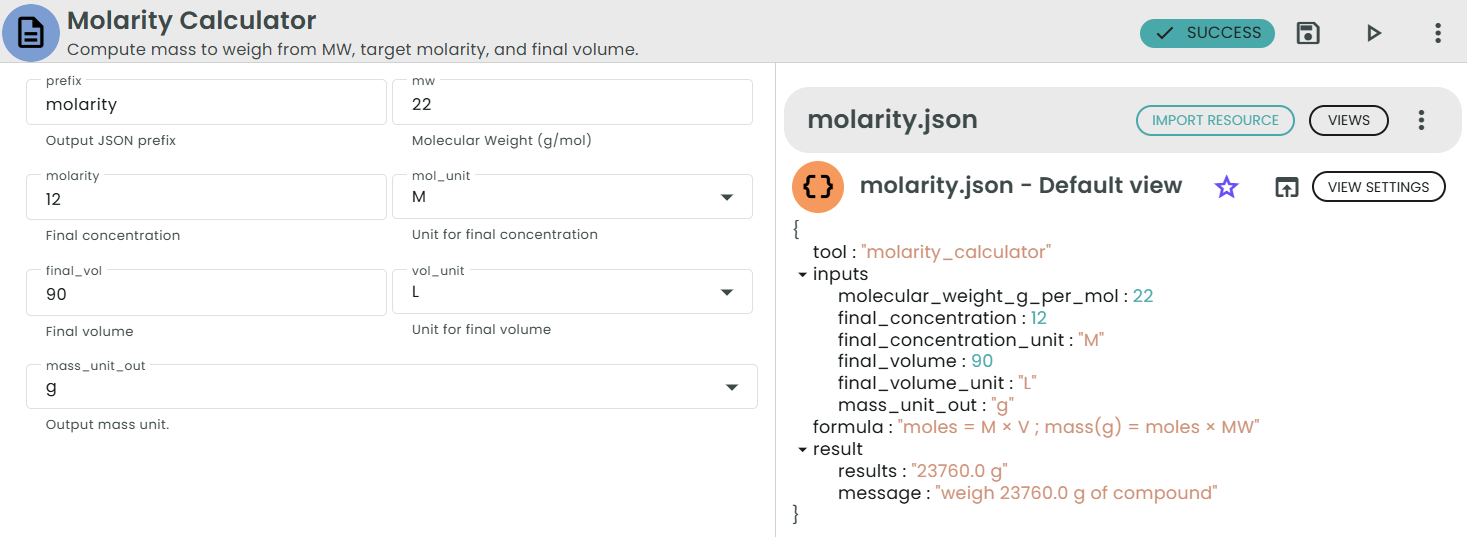
✨Molarity Calculator✨
What: Mass required to achieve a given molarity and final volume.
Why: Make solutions accurately from solid compounds.
Calculates the mass of a compound to weigh given:
• Molecular Weight (MW) in g/mol
• Desired final concentration (M) in mol/L
• Desired final volume (V) in L
Formula:
• moles (mol) = M × V
• mass (g) = moles × MW = M × V × MW
Usage:
1) Enter MW (g/mol)
2) Enter target M (e.g., 0.25 for 0.25 M)
3) Enter final volume in liters (e.g., 0.5 for 500 mL)
4) Compute mass = M × V × MW (in grams)
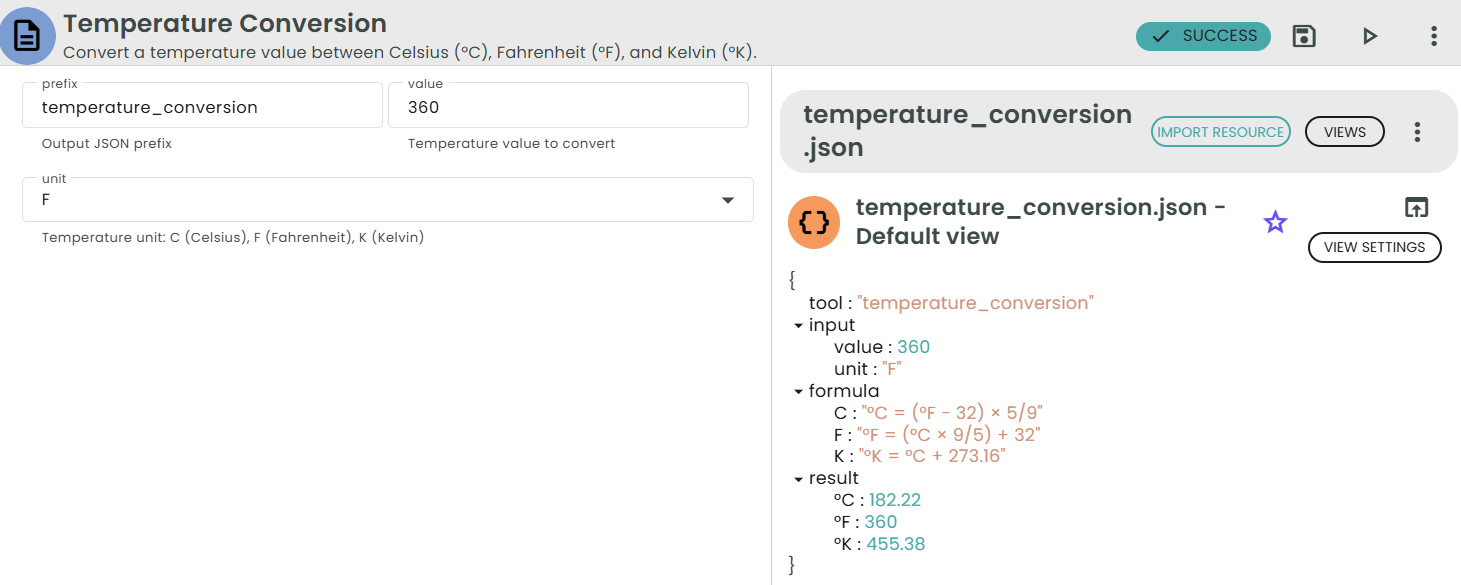
✨Temperature Conversion✨
What: Convert between °C, °F, and K.
Why: Protocols and instruments often use different temperature scales.
Converts a temperature value between:
• °C (Celsius)
• °F (Fahrenheit)
• K (Kelvin)
Formulas:
• °C = (°F − 32) × 5/9
• °F = (°C × 9/5) + 32
• K = °C + 273.16
Examples:
• 25 °C → 77 °F → 298.16 K
• 0 °F → −17.78 °C → 255.38 K
• 310 K → 36.84 °C → 98.33 °F