Introduction
The purpose of this article is to present how to work with digital twins of cellular metabolism using COBRApy, the most widely used Python package for constraint-based modeling. We will see how to load and visualize a metabolic model and how to use COBRApy to predict cell growth. We will use Constellab to perform these steps.
We start from an existing genome-scale metabolic models. We use the well known e_coli_core model of Escherichia coli str. K-12 substr. MG1655. This model was obtained from the BiGG database and published by Orth et al, EcoSal Plus 2010.
The E. coli core model
As described by Orth and co-authors, the core E. coli model is a small-scale model that can be used for educational purposes. It is meant to be used by senior undergraduate and first-year graduate students learning about constraint-based modeling and systems biology. This model has enough reactions and pathways to enable interesting and insightful calculations, but it is also simple enough that the results of such calculations can be understood easily.
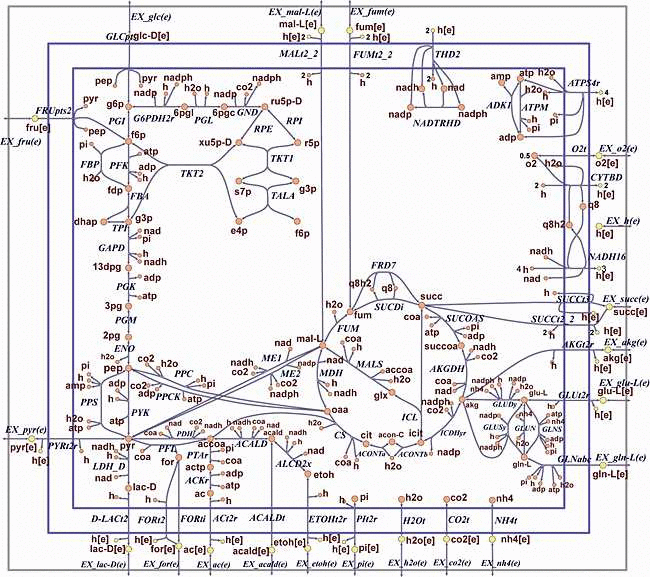
Preparation
In Constellab, create a new scenario and select a "Conda env agent" process. In your Constellab agent environment, you need to add:
channels:
- conda-forge
dependencies:
- cobra==0.29.1
Then you can add the following code of the tutoriel in the "Python code snippet" section.
Model loading and visualisation
COBRApy can load models stored in JSON, SBML, or YAML formats. Here we will load the e.coli model :
from cobra.io import load_model
ecoli_json_aerobic = load_model("textbook")
Predicting cell growth
Now that the model has been loaded, it is possible to build a digital twin and apply an Flux Balance Analysis (FBA). In the article, they model a few conditions, here we will reproduce some.
By default, the objective of the E. coli core model is the biomass reaction.
Under aerobic conditions
The model provided in BIGG is already in aerobic conditions (lower bound of "EX_glc__D_e" at = -10 and lower bound of "EX_o2_e" at -1000). If the objective function to maximize is the Biomass, then the article found a value of 0.87 h-1.
If we run the experiment in Constellab using COBRApy, we found the same value:
linear_reaction_coefficients(ecoli_json_aerobic)
aerobic_objective = ecoli_json_aerobic.optimize().objective_value
Under anaerobic conditions
Then we modify the model provided in the input to simulate anaerobic conditions.
We have made the following changes:
- Keep the lower bound of "EX_glc__D_e" at -10
- Set the lower bound of "EX_o2_e" to 0
If the objective function to maximize is the Biomass, then the article found a value of 0.21 h-1; we found the same value in Constellab with COBRApy:
ecoli_json_anaerobic.reactions.get_by_id("EX_glc__D_e").lower_bound = -10
ecoli_json_anaerobic.reactions.get_by_id("EX_o2_e").lower_bound = 0
linear_reaction_coefficients(ecoli_json_anaerobic)
anaerobic_objective = ecoli_json_anaerobic.optimize().objective_value
Changing the objective function by ATPM
Here, we will set the lower bound of "EX_glc__D_e" to -1 (The model is provided with the story, under the name "e_coli_core_ATPM.json".) and change the objective function to maximum ATP yield (reaction "ATPM").
The maximum yield of ATPM given in the article is 17.5. This value is also found in Constellab with COBRApy:
ecoli_json_atpm.reactions.get_by_id("EX_glc__D_e").lower_bound = -1
ecoli_json_atpm.objective = "ATPM"
linear_reaction_coefficients(ecoli_json_atpm)
atpm_objective = ecoli_json_atpm.optimize().objective_value
atpm_flux = ecoli_json_atpm.optimize().fluxes.ATPM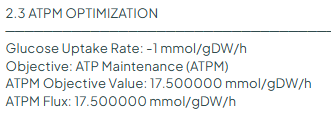
Conclusion
This pipeline demonstrates how to use COBRApy in Constellab to simulate digital twins of cellular metabolism from genome-scale models.
This workflow can be applied to any organism for which a GEM model is available. COBRApy is reliable, reproducible, and widely accepted in systems biology research.
Find here the agent related to this story where you can find the whole code.

Comments (0)
Write a comment