Bread wheat (Triticum aestivum L.) is a vital crop with a genome size that ranks among the largest ever assembled at a reference-quality level. This hexaploid crop, consisting of three subgenomes (A, B, and D), comprises approximately 15 gigabases (Gb) of DNA, with a staggering 85% composed of transposable elements (TEs). While wheat genetic diversity has predominantly been studied in terms of genes, little is known about the extent of genomic variability affecting TEs, their transposition rates, and the impact of polyploidy. However, recent advancements in genome sequencing have paved the way for a deeper exploration of these intriguing aspects.
A Multidimensional Study:
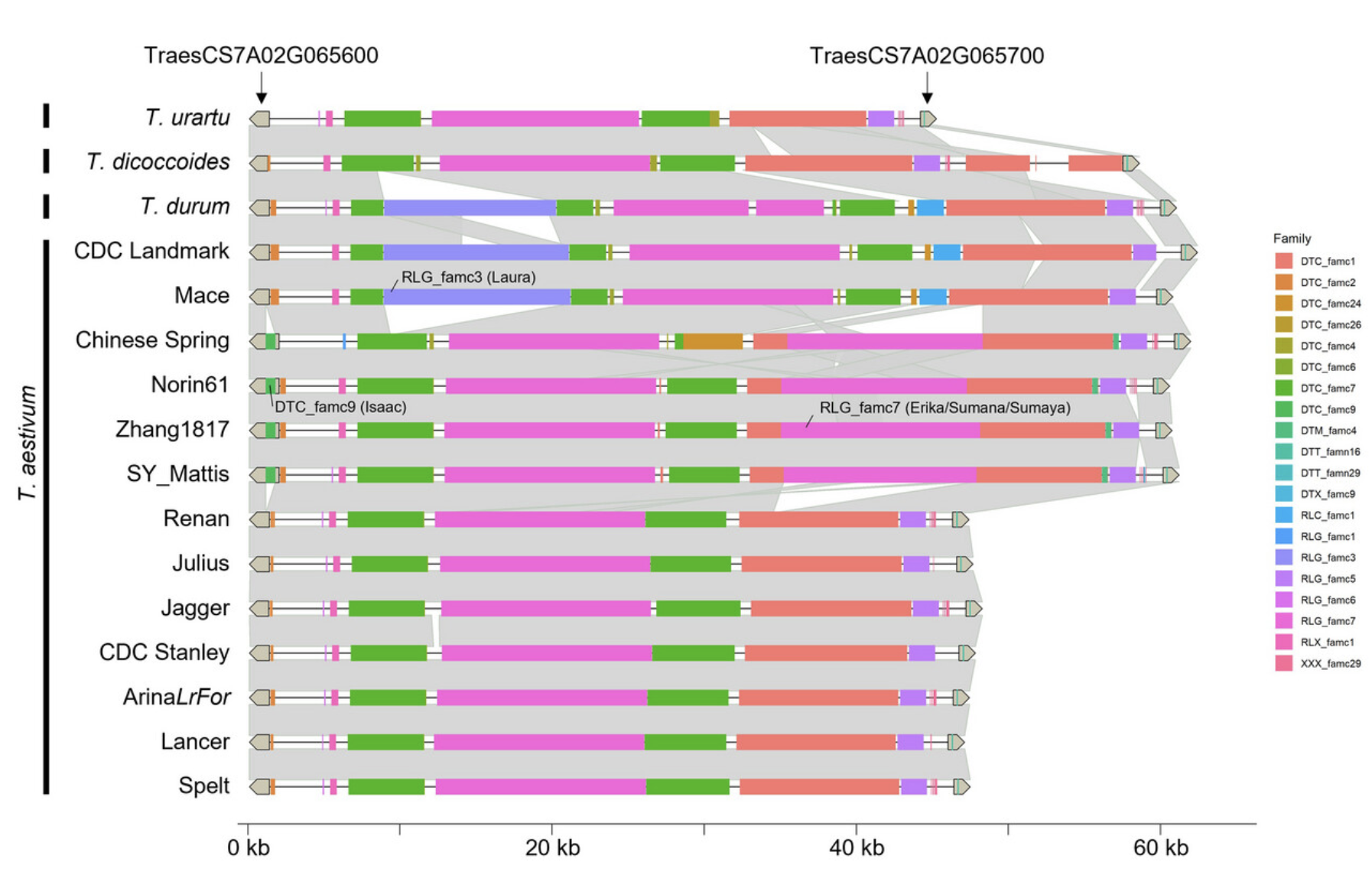
In this study published in The Plant Genome (Click Here), INRAE (Frederic Choulet’s lab – Clermont Ferrand) and Gencovery (Romain De Oliveira, PhD) researchers undertook the task of computing base pair-resolved, gene-anchored, whole-genome alignments of A, B, and D lineages at different ploidy levels to estimate the variability that affects the TE space in wheat. To achieve this, they utilized assembled genomes of thirteen T. aestivum cultivars (Bread Wheat, 6x = AABBDD), along with single genomes for Triticum durum (Pasta Wheat, 4x = AABB), Triticum dicoccoides (Wild Wheat, 4x = AABB), Triticum urartu (Wild Wheat, 2x = AA), and Aegilops tauschii (Wild Wheat, 2x = DD).
Revealing TE Variability:
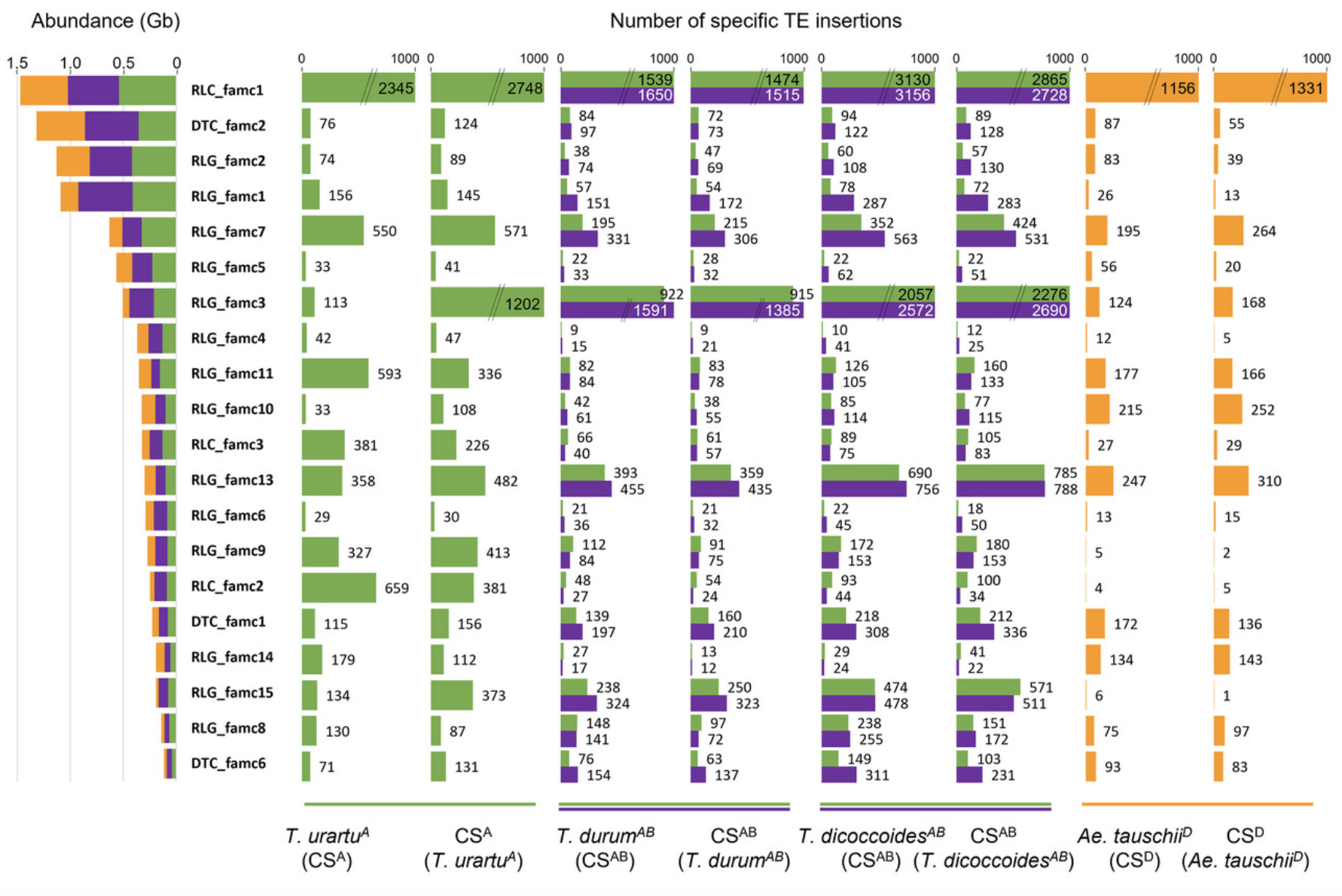
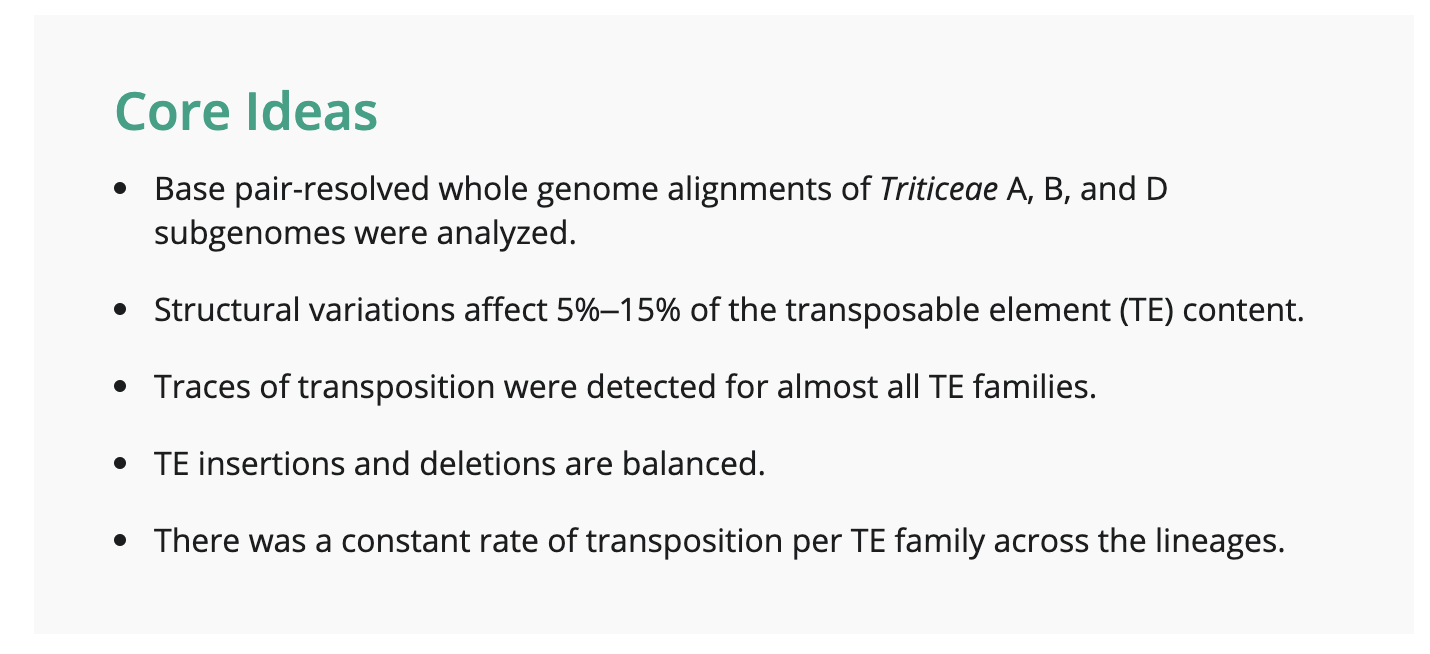
The findings of the study shed light on the previously unexplored world of TE variability in wheat. It was observed that between 5% and 34% of the TE fraction exhibited variability, depending on the species divergence. Remarkably, the researchers detected between 400 and 13,000 novel TE insertions per subgenome, highlighting the dynamic nature of these genetic elements.
Lineage-Specific Insights:
One of the interesting results from this study was the identification of lineage-specific insertions for nearly all TE families in diploid, tetraploid, and hexaploid wheat. These lineage-specific insertions provide valuable insights into the evolutionary history and genetic diversity of wheat and its wild relatives. Furthermore, these findings challenge the prevailing notion of wheat TE dynamics and support a more balanced model of evolution, suggesting an equilibrium between TE insertions and removals.
No Transposition Bursts or Polyploidization Boost:
Contrary to previous assumptions, the study found no evidence of a burst of transposition associated with polyploidization events in wheat. This challenges the conventional understanding that polyploidy triggers an increase in TE transposition rates. The absence of such transposition bursts suggests a more nuanced and balanced relationship between polyploidization and TE dynamics in wheat.
Implications and Future Directions:
This study opens new avenues for further research on the role of TEs in wheat evolution and its genetic diversity. Understanding the dynamics and impact of TEs in the wheat genome is crucial for crop improvement programs and the development of more resilient and productive wheat varieties.
Conclusion:
By delving into the hidden world of wheat genomics, this study has unravelled the intriguing variability of transposable elements and challenged prevailing assumptions about their dynamics. The discovery of lineage-specific insertions and the absence of transposition bursts during polyploidization highlight the need for a more comprehensive understanding of TE dynamics in wheat. As researchers continue to explore this complex landscape, these insights will pave the way for advancements in wheat breeding and contribute to the sustainable production of this vital crop in the face of global challenges.
Direct link to the study: https://doi.org/10.1002/tpg2.20347

Comments (0)
Write a comment