Introduction
Do you have a metabolic model and want to improve the annotation of metabolites and reactions?
Metabolic models are sometimes lacking in annotation, particularly at the level of identifiers in recognised databases (BiGG, Rhea, ChEBI). For the model to be better recognised by Constellab and for the experiments to work optimally, it is preferable to add these identifiers for both the metabolites and the reactions.
Follow the steps below to get a more robust model in just a few clicks!
Prerequesites
Steps to follow
- Upload your model
- Link it to the task available in the brick
- Fill parameters : you must indicate the type of identifier and the type of name of your metabolites and reactions (BiGG, literal name or other)
- Run your experiment
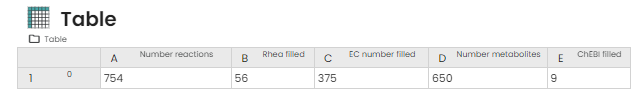
- Get your annotated model in output and a table summarising the number of identifiers added
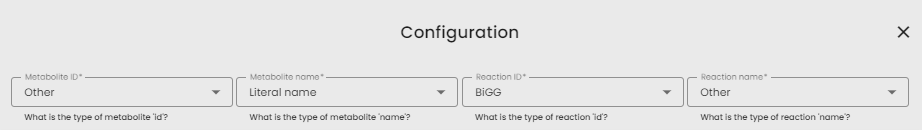
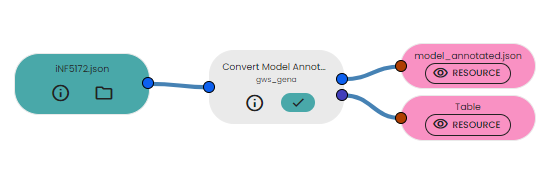
You now have a better annotated model, which is therefore more robust and will be better recognised by Constellab when you continue your experiments!

Comments (0)
Write a comment