Introduction
To delve into the study of digital twins, one requires a meticulously crafted metabolic model to facilitate in-depth analysis. Unfortunately, the current state of metabolic models lacks uniformity and robustness due to the absence of standardization. The lack of standardized protocols often leads to substantial discrepancies in quality and reliability among these models. As a result, these inconsistencies may compromise the accuracy and reliability of subsequent analyses, particularly when leveraging sophisticated tools such as Constellab.
Before integrating your model into Constellab, consider the following points to enhance the model's robustness, thus enabling more profound exploration of digital twins and facilitating more informed decision-making:
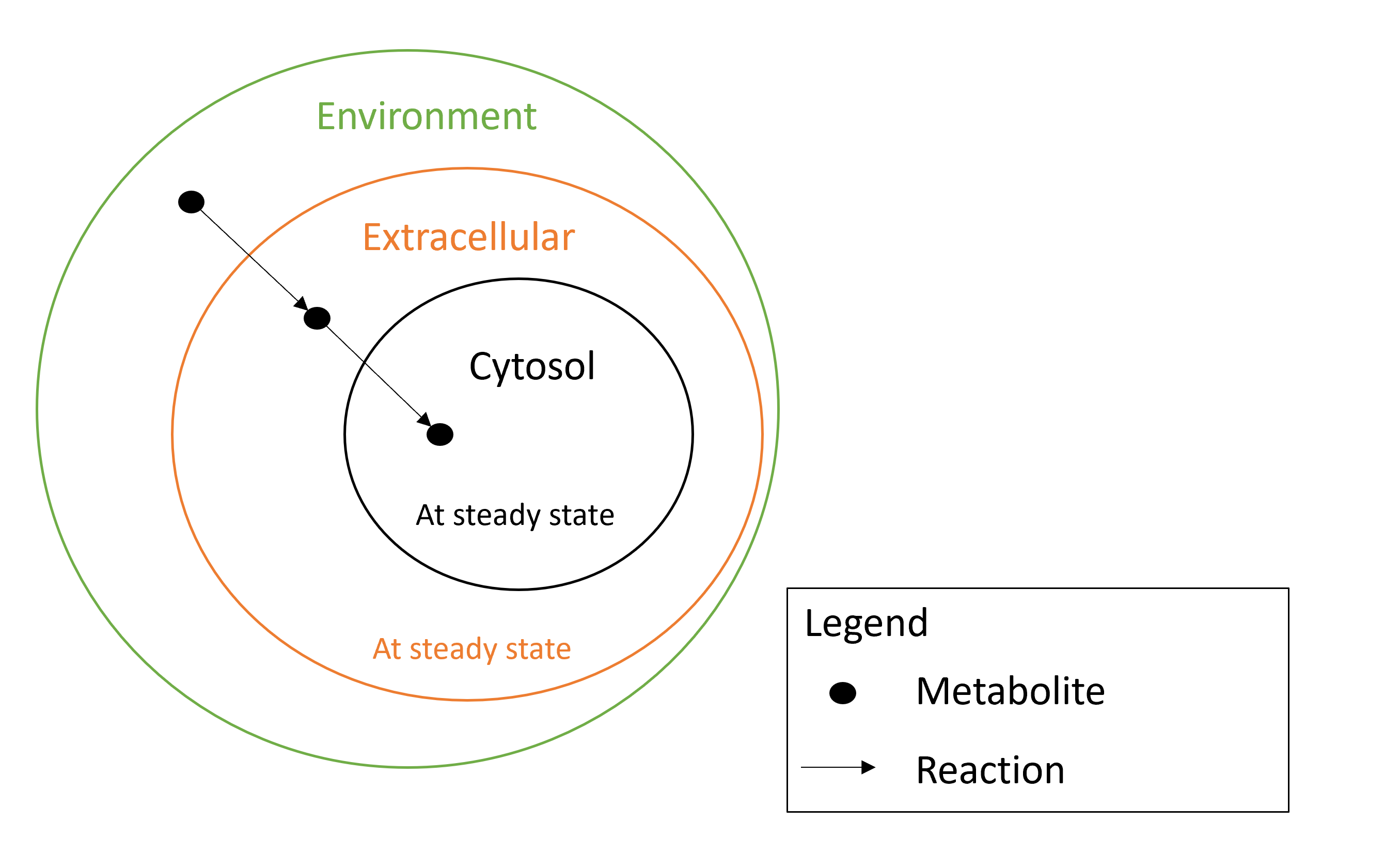
Compartment hypothesis
To model the metabolic network, several considerations are made when importing your metabolic network into Constellab. Please note the following about the compartments in order to match our representation:
- The compartment extracellular space (theoretical supernatant) (GO:0005615) is consider at steady state.
- The compartment extracellular region (environment) (GO:0005576) is consider at the non-steady state.
So, if the metabolite can be consumed, you need to set its compartment to the compartment environment (GO:0005576) and create a new metabolite in the extracellular compartment (GO:0005615) and the associated reactions. If the metabolite cannot be consumed, leave it in the extracellular compartment.
To summarize see the diagram below:
Biomass metabolite
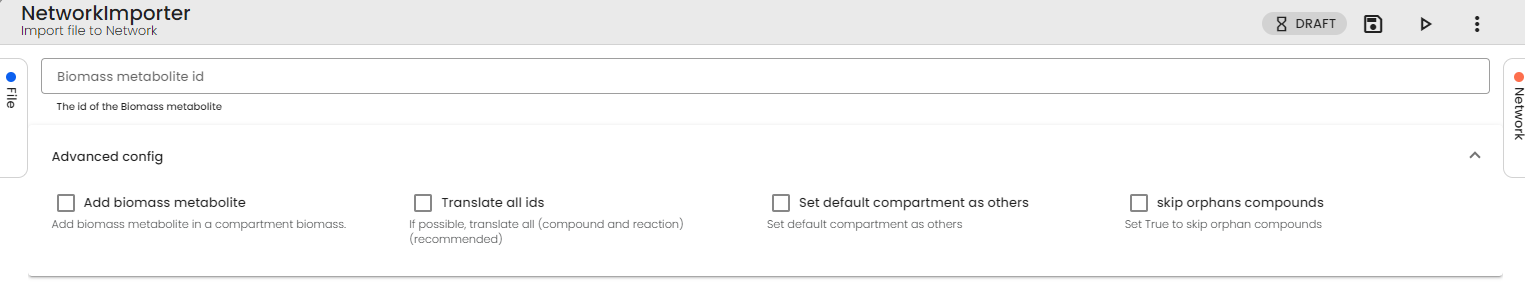
In order to predict the growth rate (e.g. using Flux Balance Analysis), your model must include a metabolite to represent the production of biomass. This metabolite must be produced by one reaction and not used as a substrate in another. Furthermore, this metabolite must be in the biomass compartment (GO:0016049).
This metabolite ID must be specified in the parameters of the Network Importer Task.
If you don't have a metabolite to represent your biomass in your model, please create one manually or set the "Add Biomass Metabolite" parameter to True. We will then add one to your model for you.
Conclusion
With a comprehensive grasp of the elements necessary for sustaining a formidable metabolic model, you're now prepared to delve into the intricacies of digital twin simulations with assurance. By placing emphasis on the meticulous curation of your model and adhering to the provided guidelines, you will have a solid foundation to explore digital twin complexities confidently. This robust model, when integrated into Constellab, empowers you to harness the tool's complete potential, facilitating profound insights and catalyzing significant breakthroughs within your specific research or application domain.

Comments (0)
Write a comment