Introduction
There are many methods for constructing genome-scale metabolic models, and many models have already been constructed and are available in public databases such as BiGG. Nevertheless, the lack of standardised annotation of metabolite names and identifiers can lead to multiple models being built but incompatible.
The aim of this story is to present a task available in Constellab that allows you to merge two metabolic networks!
Prerequesites
You need to have a least two metabolic networks.
Steps to follow
- Upload at least two metabolic networks in Constellab.
- Link them to the Task "Network merger"
- Get your extended network!
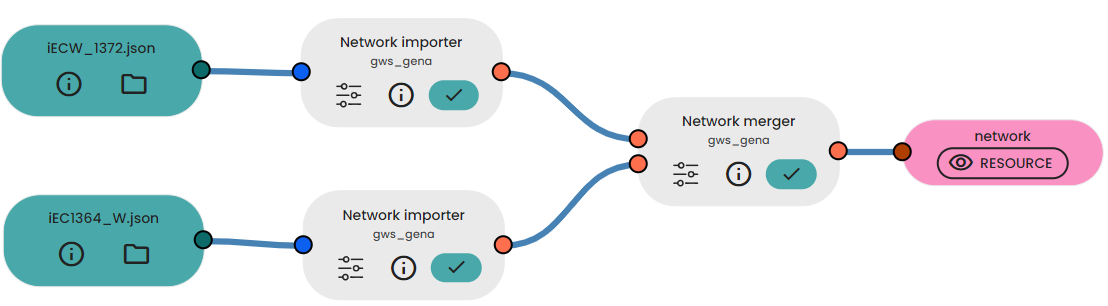
Results
The output is a merged network: the reactions of the second network that are not present in the first network are added with their associated metabolites.
For example, here we get a model with 2424 metabolites and 2935 reactions.
In the input we have a first model with 1973 metabolites and 2782 reactions and a second with 1927 metabolites and 2764 reactions. We can see that there are no duplicate reactions, so our model is well extended!
Conclusion
In conclusion, this Task is a simple Task that can be used to combine information from two different metabolic networks. This can be useful if you have two drafted networks or if you want to add your own specifications to an already published network.
You can repeat this task with other networks until you have larger networks with all the possible information.

Comments (0)
Write a comment