Introduction
The National Center for Biotechnology Information (NCBI), founded in 1988 as part of the National Library of Medicine (NLM), plays an important role in the field of biotechnology and bioinformatics. Its mission is to provide access to a vast collection of biomedical and genetic data, to promote research in these fields and to facilitate the understanding of biological processes through advanced computational tools. NCBI has become a key resource for researchers, clinicians and educators worldwide.
NCBI hosts a variety of databases covering a multitude of biomedical and genetic data types. For example, PubMed is a biomedical literature search engine containing millions of abstracts and scientific articles, providing essential access to medical and biological research.
The data available via NCBI is varied and includes not only nucleotide and protein sequences, but also information on macromolecular structures, gene expression studies, genetic variations and biomolecular interactions.
Prerequesites
- An acces to Constellab and to a digital lab
- The brick “gws_biota” (version >= 0.7.4)
Steps to follow
- Create a new experiment
- Import the “Request NCBI” task, write your request : for example “actin” and choose your database of interest (pubmed, Gene, Geo DataSet, popset, geo profiles, protein, clinvar).
- Import the “Download NCBI proteome file” task if you want to quickly download the proteome of a particular species.
Results
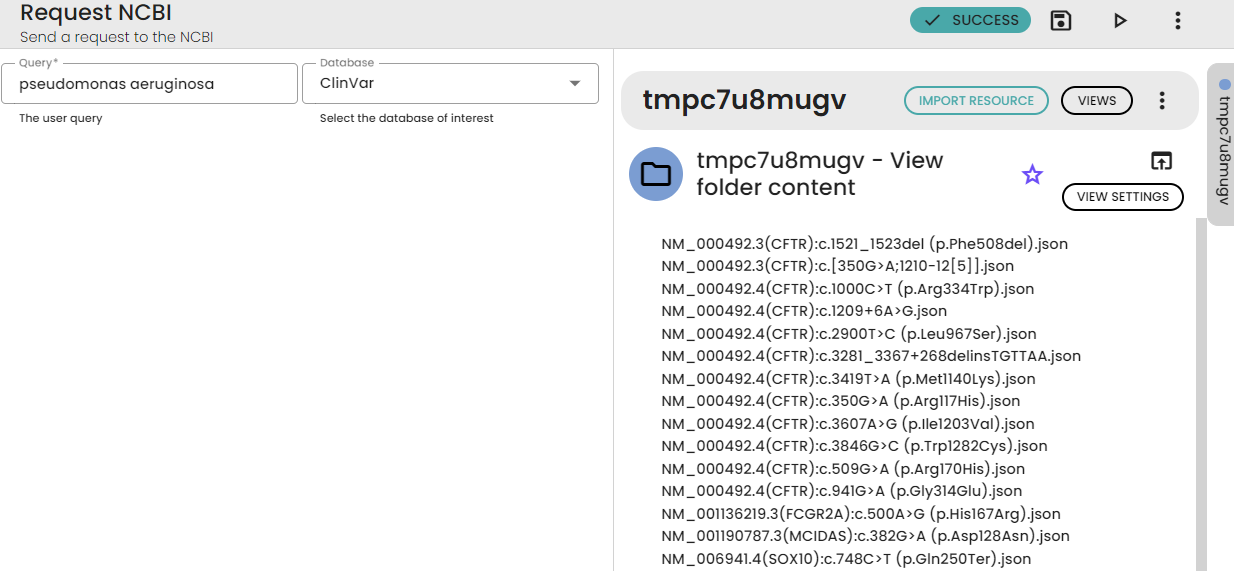
With the “Request NCBI” task, for example, you write “pseudomonas aeruginosa” as a query and choose the “ClinVar” database to obtain all the information on genetic variations and their relationship with human health. When you perform a query in ClinVar for “Pseudomonas aeruginosa”, you'll get specific information related to this bacterium in terms of its interactions or impacts on human health. If you choose the “pubmed” database, you'll get the references and abstracts of articles related to your query. If you choose the “Geo DataSet” database, you'll get datasets related to gene expression associated with that bacterium (e.g. from high-throughput gene expression profiling experiments, such as microarrays, RNA-Seq, and other functional genomics techniques). Etc.
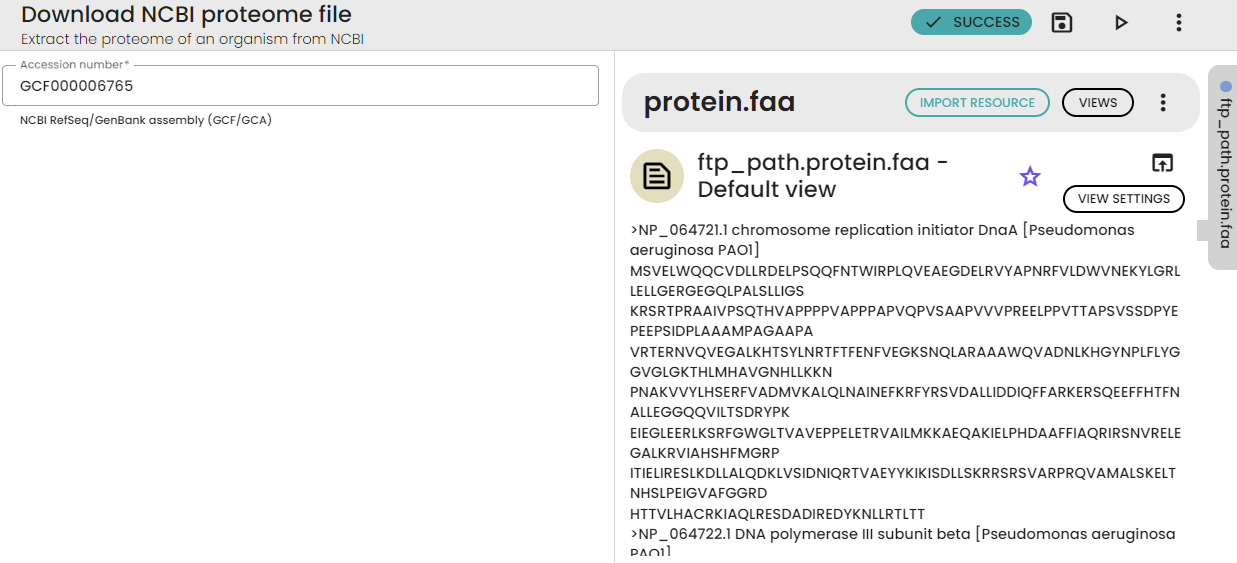
The “Download NCBI proteome file” task asks for the GCF or GCA identifier of the species of interest, e.g. the identifier for pseudomonas aeruginosa is “GCF000006765”.
Conclusion
In conclusion, it's important to note that query results are limited to 100 000 results per search. This underlines the need to perform targeted rather than broad queries to obtain relevant, usable results. For example, a general query such as “actin” will generate a considerable number of results in all NCBI databases, due to the abundance of studies on this essential and widely studied protein. On the other hand, a more specific query like “oprf” - a less common protein - will produce a much smaller number of results, allowing a more precise and maneuverable exploration of the available data.

Comments (0)
Write a comment