Introduction
The purpose of this article is to present how to use digital twins of cellular metabolism in Constellab. We will see how to visualize a model in Constellab and how to predict cell growth.
We start from an existing genome-scale metabolic models. We use the well known e_coli_core model of Escherichia coli str. K-12 substr. MG1655. This model was obtained from the BiGG database and published by Orth et al, EcoSal Plus 2010.
The E. coli core model
As described by Orth and co-authors, the core E. coli model is a small-scale model that can be used for educational purposes. It is meant to be used by senior undergraduate and first-year graduate students learning about constraint-based modeling and systems biology. This model has enough reactions and pathways to enable interesting and insightful calculations, but it is also simple enough that the results of such calculations can be understood easily.
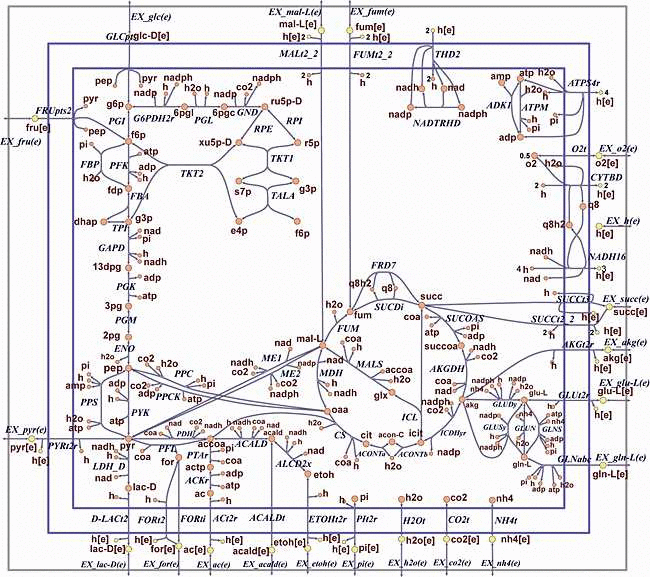
It is really usefull and is systematically used to challenge algorithms developed into Constellab.
Model loading and visualisation
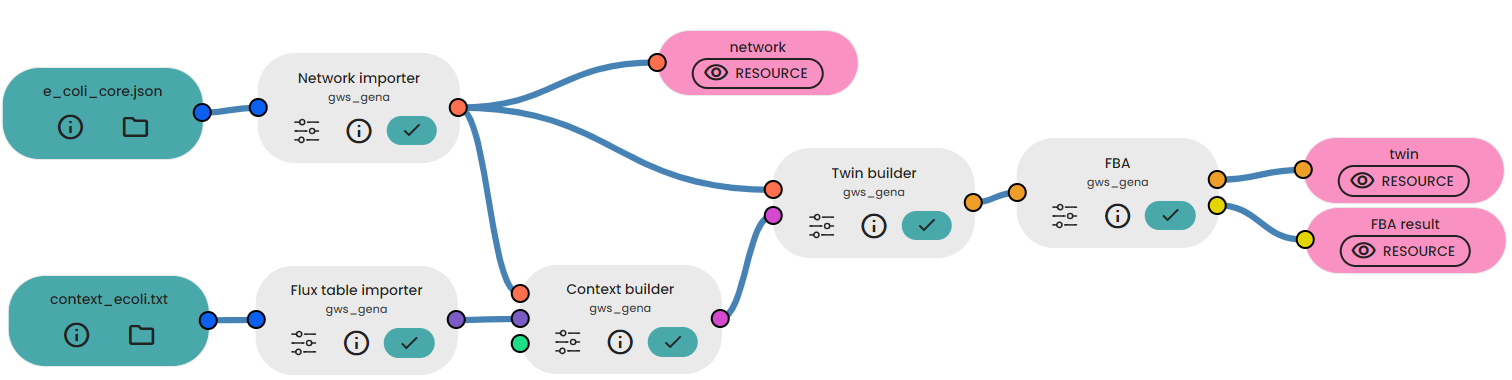
In Constellab, the model import is done via the Databox. The model is provided with the story, under the name "e_coli_core1.json".
- Go to the Databox tab
- Click on "Upload files" at the top right and select your model.
- Specify the type of your file as "NetworkFile".
- Once your file is uploaded, click on the 3 small dots at the end of the line of your resource and then on "Import resource".
- Select "Network importer" in "Select importer" and then click on "Import Resource".
And now your model is imported! To view it, click on the eye on the line of your metabolic network. A window will open, showing your entire network!
Predicting cell growth
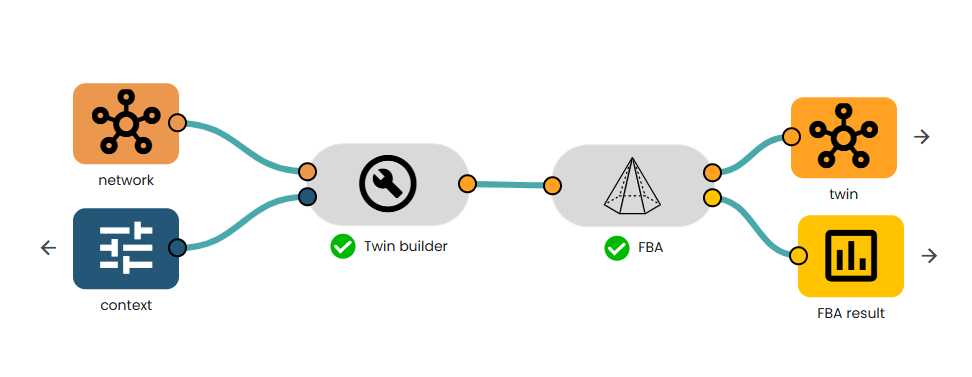
Now that the model has been imported into Constellab, it is possible to build a digital twin and apply an Flux Balance Analysis (FBA).
Steps to follow
- Upload you model e.coli and a empty context (it will be possible to add to it later if you wish).
- Link your context to the "Context importer" task and then link the output and your model to the task available in the brick.
- Then, link the twin to the "FBA" task.
- Fill parameters : you can choose to maximize or minimize the biomass or other fluxes, you need to specify the solver, if you want to relax QSSA, among others.
- Run your experiment
- Get your twin annotated, and the estimates fluxes!
In the article, they model a few conditions, here we will reproduce some to show the accuracy of the FBA in Constellab.
For your information, the simulations were also run on a notebook using COBRApy. You can find this file ("FBA_e_coli_with_cobrapy.ipynb") attached to the story.
You can also run COBRApy directly in Constellab, using agents for example. Find here the agent related to this story.
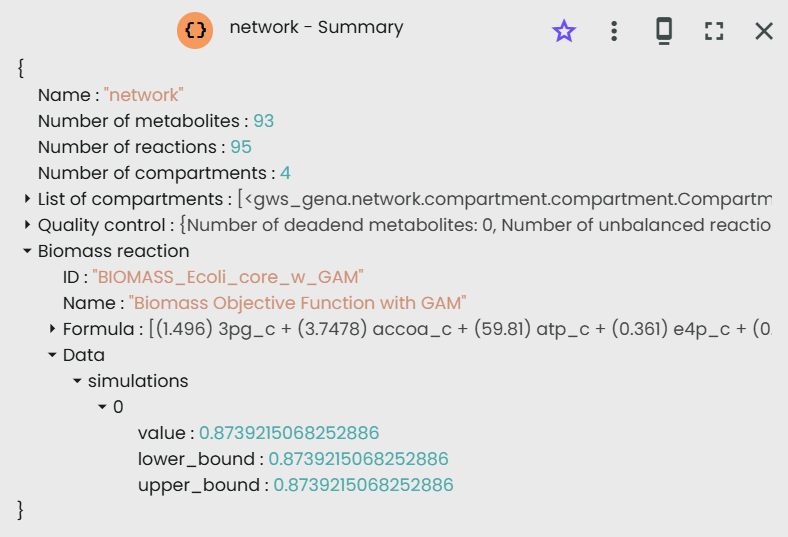
Under aerobic conditions
The model provided in BIGG is already in aerobic conditions (lower bound of "EX_glc__D_e" at = -10 and lower bound of "EX_o2_e" at -1000). If the objective function to maximize is the Biomass, then the article found a value of 0.87 h-1.
If we run the experiment in Constellab as described previously, we found the same value: by clicking on "twin" in the output resources, then on "summary", you can find the estimated value of biomass production.
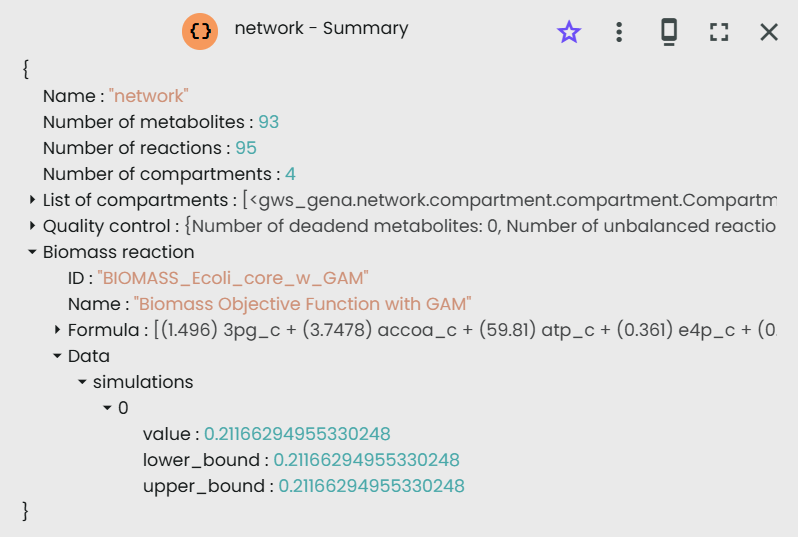
Under anaerobic conditions
Then we modify the model provided in the input to simulate anaerobic conditions.
We have made the following changes:
- Keep the lower bound of "EX_glc__D_e" at -10
- Set the lower bound of "EX_o2_e" to 0
The model is provided with the story, under the name "e_coli_core_anaerobic.json".
If the objective function to maximize is the Biomass, then the article found a value of 0.21 h-1; we found the same value in Constellab:
Changing the objective function by ATPM
Here, we will set the lower bound of "EX_glc__D_e" to -1 (The model is provided with the story, under the name "e_coli_core_ATPM.json".) and change the objective function to maximum ATP yield (reaction "ATPM"). To do this, we need to set the parameters of the FBA task as follows:
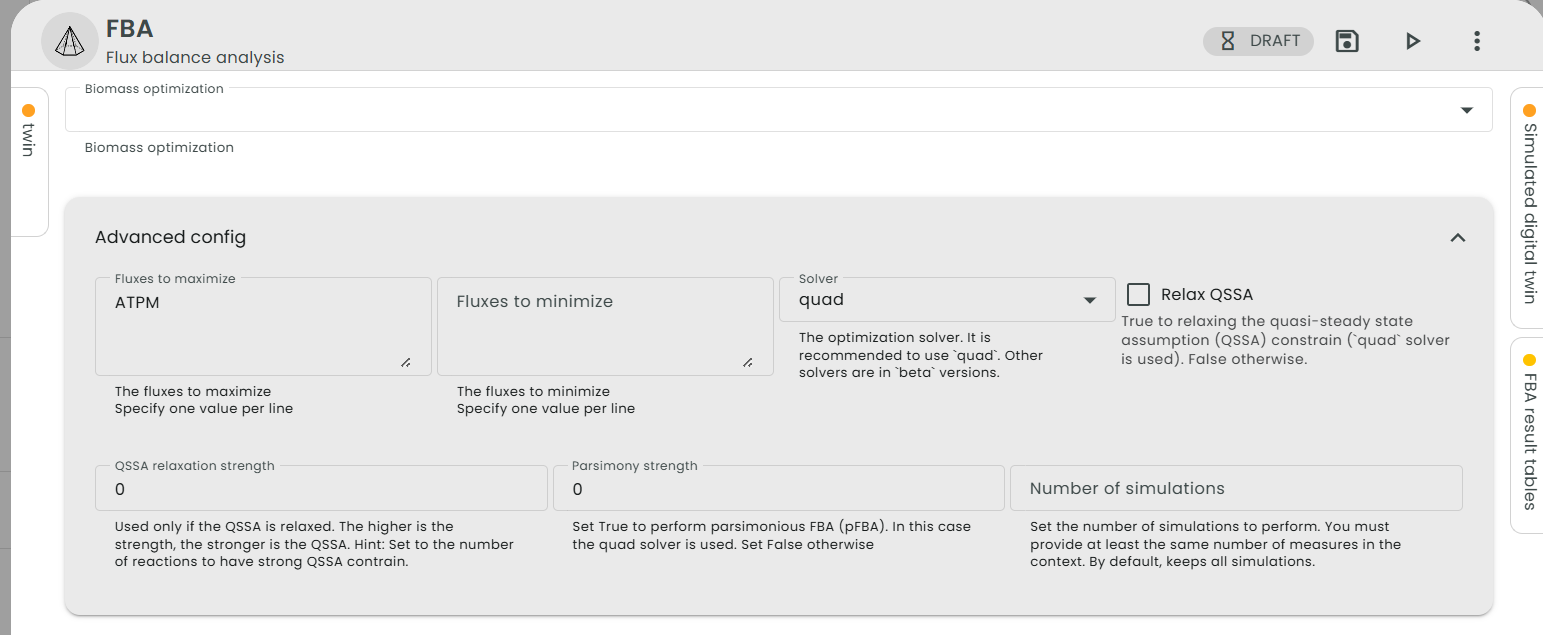
The maximum yield of ATPM given in the article is 17.5. This value is also found by Constellab. To retrieve it, you need to go on the Flux Table and search for the reaction "ATPM".
In view of the good reproducibility of the results in the article, we have proved that the FBA implemented in Constellab can be used in your projects.
Performing knock-out analyses
Depending on what you want to analyse, you can perform a knock-out analyse. This makes it possible to delete genes to study the effects of deleting the target gene on cellular processes, molecular interactions or the development of the organism.
Conclusion
This pipeline allows you to simulate digital twins of cellular metabolism from GEM models in Constellab.
This tutorial has allowed us to show that our FBA Task retrieves the results of the reference article on e.coli.
So this pipeline can be used with any organism to obtain simulated fluxes.

Comments (0)
Write a comment