Introduction
Simulation of gene knockout involves using computational techniques to predict the effects of inactivating genes within biological systems. By selectively deactivating specific genes, researchers can simulate the effects on cellular or organismal behavior, offering key insights into gene roles, regulatory pathways, and potential therapeutic targets.
These simulations employ mathematical models such as constraint-based metabolic models or boolean networks to simulate the modified cellular phenotype following gene knockout. Through these simulations, researchers can probe how gene knockout influences cellular metabolism, contributing to the identification of essential genes, understanding disease mechanisms, and devising strategies for genetic engineering or drug discovery. Gene knockout simulations play an essential role in systems biology, providing a powerful tool for hypothesis formation, experimental design, and improving our understanding of complex biological systems.
We will reproduce in Constellab the pipeline published in this paper which uses the COBRA toolbox.
Prerequesites
First, we import the e. coli core model, modified to meet the specifications of the article. So we set the lower bounds of the EX_o2_e reaction at 0 and the EX_glc__D_e reaction at -20.
Then we want to knockout these following reactions: ALCD2x, FUM, GLUDy, ME2, PYK to optimize the lactate secretion. So we need to import these reactions, here we will use an Entity ID table.
All essential files for this model are attached to this story.
Steps to follow
- Import the metabolic network, an empty context and the Entity ID Table with the multiples reactions to knockout.
- Link them to the Twin Builder and KOA tasks as shown in the pipeline below.
- In the KOA task, maximize the biomass.
- Link the KOA result to the "KOA Result Extractor" Task. Set "network_BIOMASS_Ecoli_core_w_GAM" to the params to extract the biomass flux.
- Get the estimated fluxes and especially the estimated biomass flux!
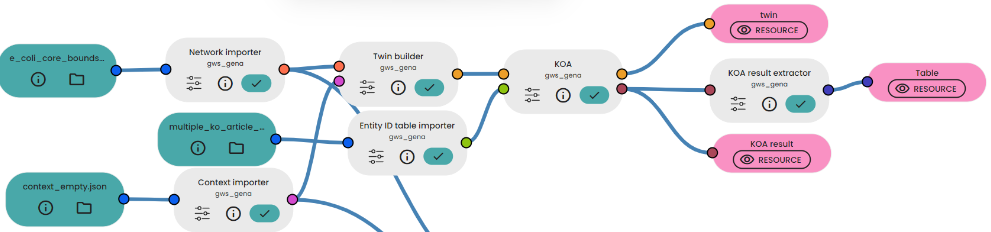
Results
After running the pipeline we get the following results:

Thus, the estimated biomass flux when we knock out the reactions ALCD2x, FUM, GLUDy, ME2, PYK is 0.142 h^-1.
The results of the article are presented below:
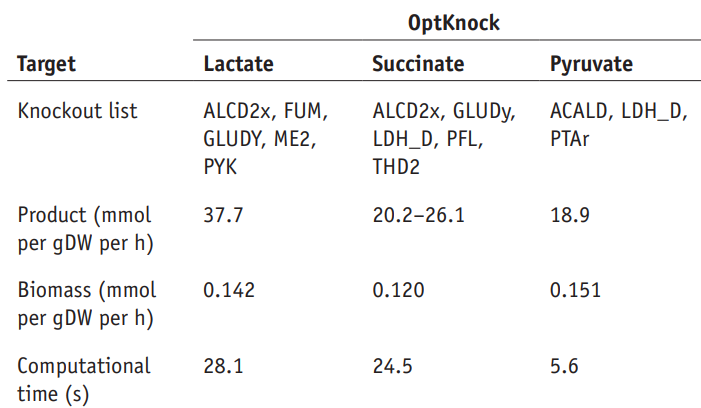
The results obtained with Constellab are therefore consistent with the published results.
Conclusion
In conclusion, the pipeline presented here in Constellab allows you to reproduce the results of the article, which means that our pipeline is robust and can be used for your own analysis!
References
- Schellenberger J, Que R, Fleming RM, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S, Kang J, Hyduke DR, Palsson BØ. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat Protoc. 2011 Aug 4;6(9):1290-307. doi: 10.1038/nprot.2011.308. PMID: 21886097; PMCID: PMC3319681.

Comments (0)
Write a comment