Short description
uBiome provides core features for microbiome sequencing data analysis.
How that works?
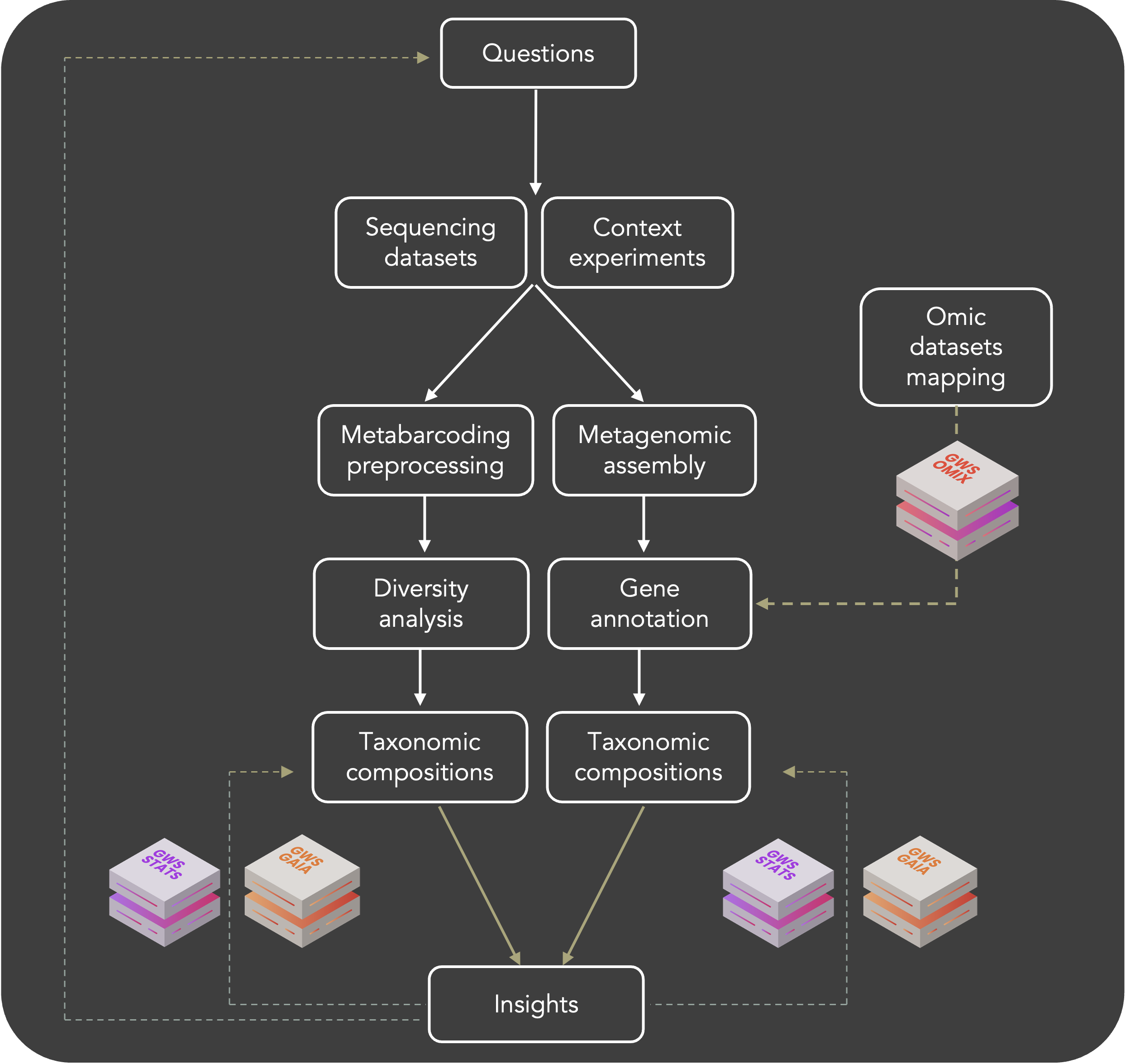
uBiome brick allows you to process your microbiome data of interest to answer your biological questions. Our pipeline was designed to process metabarcoding sequencing (rRNA and ITS) and shotgun sequencing.
Briefly, for metabarcoding analysis qiime2 tasks has been implemented and are available from sequencing quality check to differential analysis.
For shotgun sequencing, dataset metagenome assembly and further characterisations are possible (diversity assessment, genome annotation, functional assignments...).
Applications and benefits
- Assess samples taxonomic composition (microbiota, bioreactor, environemental...)
- Understand host-pathogen interactions
- Optimize organisms and culture media
- Monitoring microbial communities
Input data
- Sequencing datasets (Metabarcoding, Shotgun)
required
Timing
- Variable analysis time depending on the size of the data, the type of analysis and the specification of the machine used to analyse the data
Support
- Gencovery team support (Assembly, clustering, analysis)
- Gencovery ecosystem support (we make the connection with the right expert)
Intellectual property
- You keep IP on your proprietary data, and any related models or insights generated in Constellab™.
- You keep IP on your custom pipelines, know-how, and bricks created in Constellab™.

