🔍 Introduction
EggNOG-mapper is a powerful tool for the fast and precise functional annotation of novel sequences such as proteins, coding sequences (CDS), genomes, and metagenomes. It leverages precomputed orthologous groups (OGs) and phylogenies from the EggNOG database (eggnog5.embl.de) to ensure annotations are transferred only from fine grained orthologs, avoiding misleading annotations from close paralogs.
This tool significantly improves annotation accuracy compared to traditional homology-based tools like BLAST, making it a preferred solution for researchers working on novel genomes, transcriptomes, or metagenomic datasets.
EggNOG-mapper has been wrapped into a Constellab Task, enabling easy integration into bioinformatics workflows with reproducibility and scalability.
🧰 Prerequisites
- Access to Constellab and a valid Digital Lab environment
- Installed bricks:
gws_omixversion ≥ 0.11.6 - Input files: A FASTA file of proteins or nucleotide sequences (CDS/genome/metagenome/proteins)
🧪 Use Case Steps
I. Functional Annotation with EggNOG
- Import your FASTA file into Constellab.
- Link it to the Task:
"eggNOG Mapper". - Configure Parameters:
itype: Choose your sequence type (proteins,CDS,genome, ormetagenome).cpus: Number of CPU threads to allocate (default: 25). - Run the Task: The task automatically:
Checks and downloads required EggNOG databases.
Runs
emapper.pyusing DIAMOND for alignment. Extracts and cleans the final annotation file.
Output
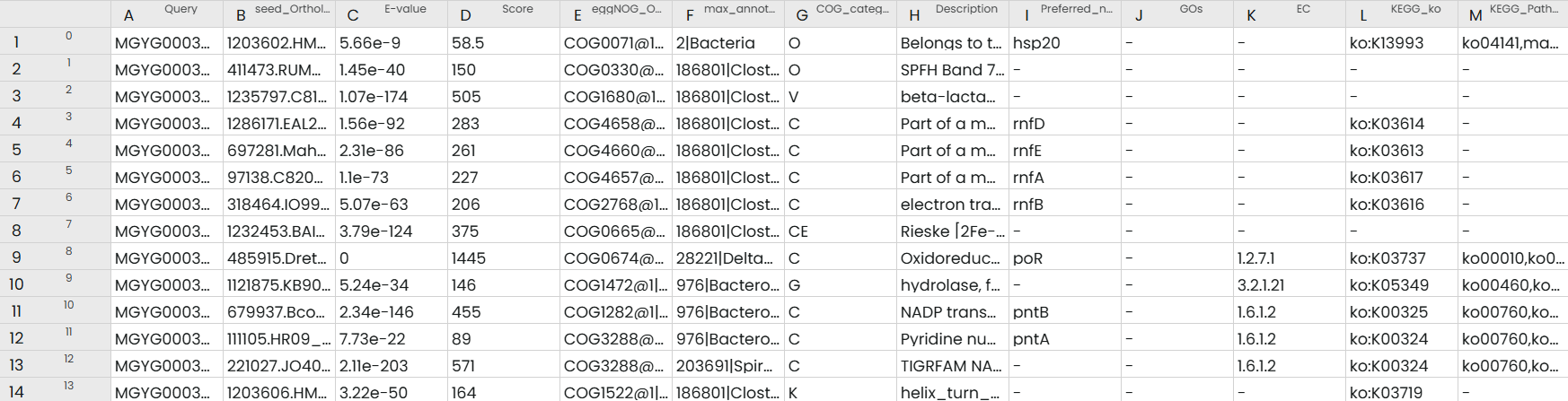
The task produces a clean, standardized annotation.tsv file containing:
- Ortholog assignments
- Functional categories (COG, GO, KEGG, etc.)
- Predicted functions and pathway mappings for each input sequence
This table can then be used for downstream tasks such as enrichment analysis or KEGG pathway.
✅ Validation with Publicly Available Dataset
To validate the workflow, the file MGYG000307600.faa was downloaded from the mgnify database via the following link: https://ftp.ebi.ac.uk/pub/databases/metagenomics/mgnify_genomes/chicken-gut/v1.0.1/species_catalogue/MGYG0003076/MGYG000307600/genome/.
This FASTA file was used as input for the EggNOG-mapper pipeline. After running the script, the resulting annotation file was compared to the functional annotations published in the mgnify database.
The results were consistent and comparable, demonstrating the reliability and accuracy of the pipeline for functional annotation.

Comments (0)
Write a comment