Introduction
MultiQC is a modular tool to aggregate results from bioinformatics analyses across many samples into a single report. It simplifies the analysis of large-scale sequencing data by automatically generating a comprehensive QC report. MultiQC takes input from various bioinformatics tools, such as alignment, variant calling, transcriptomics, and ChIP-seq pipelines, and combines their QC results into a single HTML report
Main functions of MultiQC :
- Aggregating QC Metrics : MultiQC collects quality control metrics from various bioinformatics tools and pipelines.
- Generating QC Reports : MultiQC generates comprehensive and interactive QC reports in HTML format.
- Standardizing Results : MultiQC standardizes the presentation of QC metrics across different tools and pipelines.
- Visualizing QC Metrics: The generated MultiQC report includes interactive plots, tables, and charts that visualize the QC metrics. This visual representation helps in quickly identifying trends, patterns, and potential issues in the data.
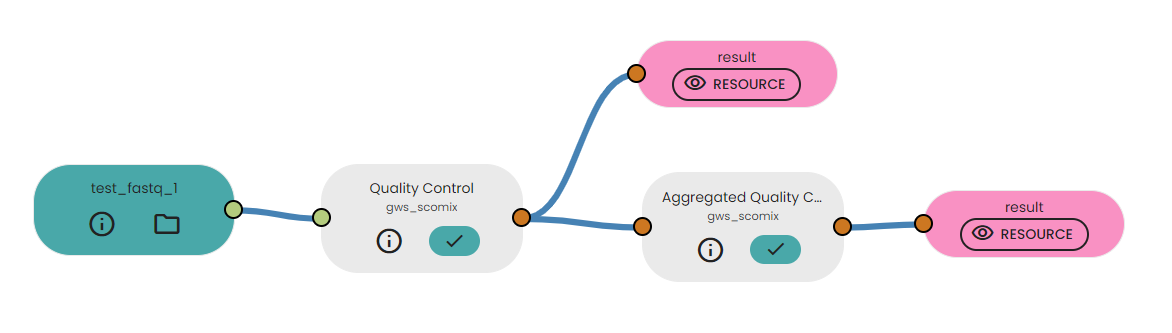
Steps to follow
Ensure that the version 0.1.2 brick is loaded
- First upload your fastqc folder to the Databox.
- Then, create a new experiment.
- Import your resource
- Link it to the task "Quality Control" available in the brick
- Link the output of task to "Aggregated Quality Control" task.
- Run your experiment.
- Get your HTML report that will provide detailed information on the quality of the input sequencing data.
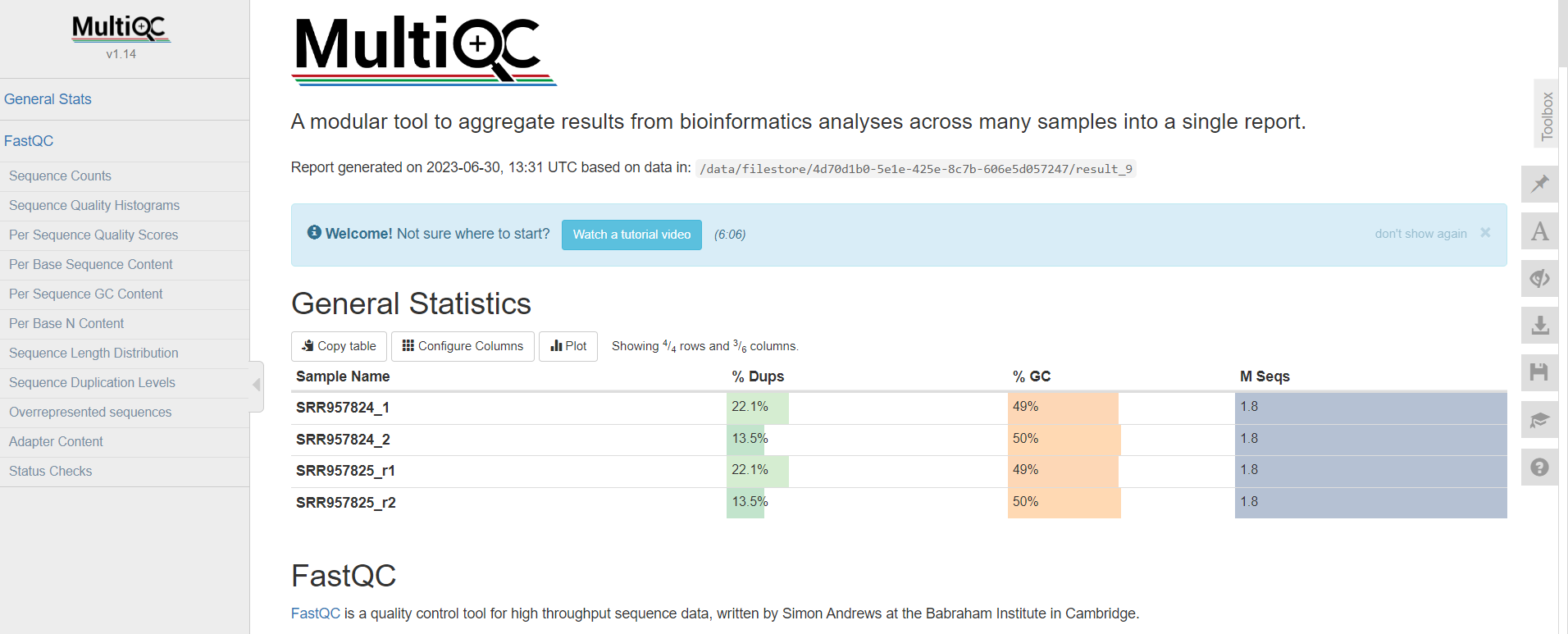
Description of output file
The report includes interactive visualizations, tables, and summary statistics that help researchers assess the quality and performance of their data analysis.

Comments (0)
Write a comment