Introduction
FastQC is a widely used and powerful quality control tool that aims to simplify the process of checking the quality of raw sequence data generated by high-throughput sequencing pipelines. It offers a modular set of analyses that allow users to quickly assess the quality of their data and identify potential issues that may need to be addressed before proceeding with downstream analyses. It is easy to use and is compatible with a variety of sequencing platforms, making it a valuable tool for researchers working with different types of sequencing data.
Main functions of FastQC :
- Providing a quick overview to tell you in which areas there may be problems
- Summary graphs and tables to quickly assess your data
- Export of results to an HTML based permanent report
Steps to follow
Ensure that the
- version 0.1.2 brick is loaded
- So first, upload your fastqc folder to the Databox.
- Then, create a new experiment.
- Import your resource
Link it to the
task available in the
- brick.
- Run your experiment
- Get your HTML report that will provide detailed information on the quality of the input sequencing data
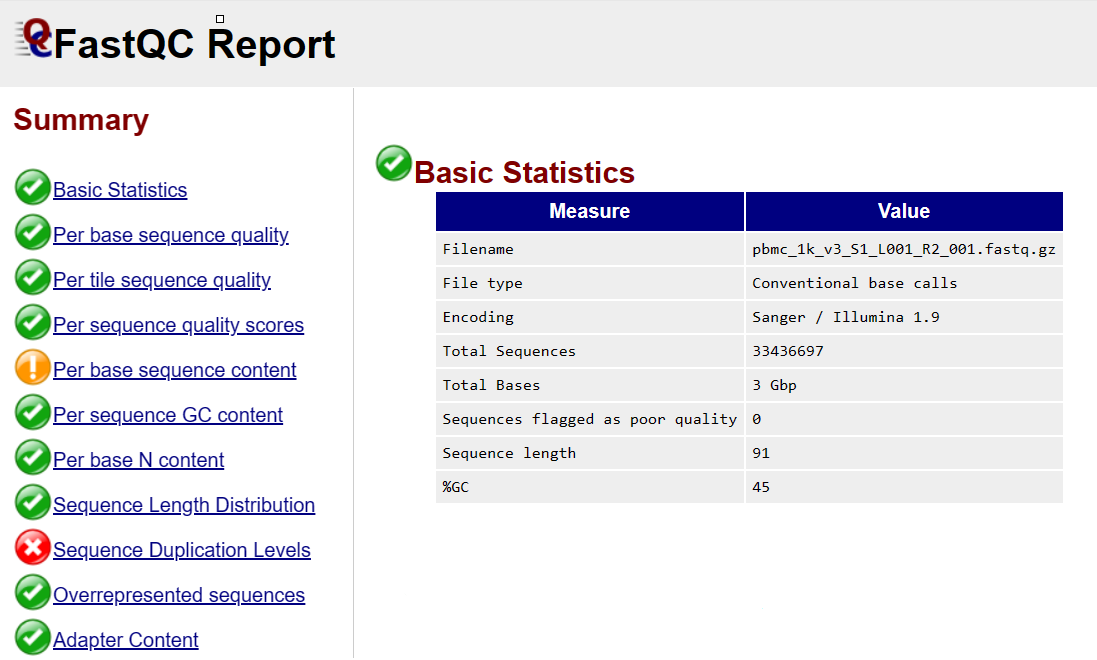
Description of output file
The reports include a variety of visualizations and statistical analyses, such as quality score plots, sequence length distribution plots, per-base sequence quality scores, per-base sequence content, GC content plots, adapter content, and overrepresented sequences, among others.

Comments (0)
Write a comment