🔍 Introduction
When you merge datasets, run downstream analyses, or build visualizations, you almost always need one canonical identifier system (e.g., Ensembl IDs or Entrez IDs). The problem: the same gene may appear as HGNC symbols, Ensembl stable IDs, Entrez Gene IDs, UniProt accessions, microarray probes, and more.
g:Profiler – g:Convert solves this by providing a central, regularly updated service that translates gene identifiers across dozens of namespaces for hundreds of organisms. This task ( OmiX – Gene_ID_conversion ) wraps g:Convert so you can normalize IDs from a table in a simple, reproducible way—no manual lookups, no stale spreadsheets.
The list of supported databases (namespaces) in g:Profiler — useful for filling your target_namespace or numeric_namespace parameters:
- Namespaces list ▶ https://biit.cs.ut.ee/gprofiler/page/namespaces-list
the link for the list of supported organisms in g:Profiler:
- Organisms list ▶ https://biit.cs.ut.ee/gprofiler/api/util/organisms_list
🧰 Prerequisites
- Access to Constellab and a valid Digital Lab environment
- Installed bricks: gws_omix ≥ 0.11.12
- An input table (CSV or TSV), such as: a DEG results file, a one-column list of identifiers, or any table that contains a column with gene IDs.
🧪 What the task does (Steps)
This task performs identifier harmonization on your gene IDs using g:Profiler g:Convert:
- Reads your table (CSV/TSV) and picks the ID columnIf you leave
id_columnempty, the first column is used automatically. - Queries g:Convert for your organism and target namespace
Examples of target namespaces:
ENSG,ENTREZGENE,ENSGACC,UNIPROT, etc. - Handles “bare numbers” correctly with the Numeric IDs treated as option
If your inputs are just numbers (e.g.,
2167,2289, …) and you want them treated as Entrez Gene IDs, set:target_namespace = ENTREZGENE(what you want to obtain)numeric_namespace = ENTREZGENE_ACC(how to interpret the numeric inputs) - Writes a clean table with the mapping result:
incoming,converted,name,description
📥 Inputs
- File: CSV or TSV containing your gene identifiers Mixed identifiers are fine; g:Profiler handles a lot of cases.
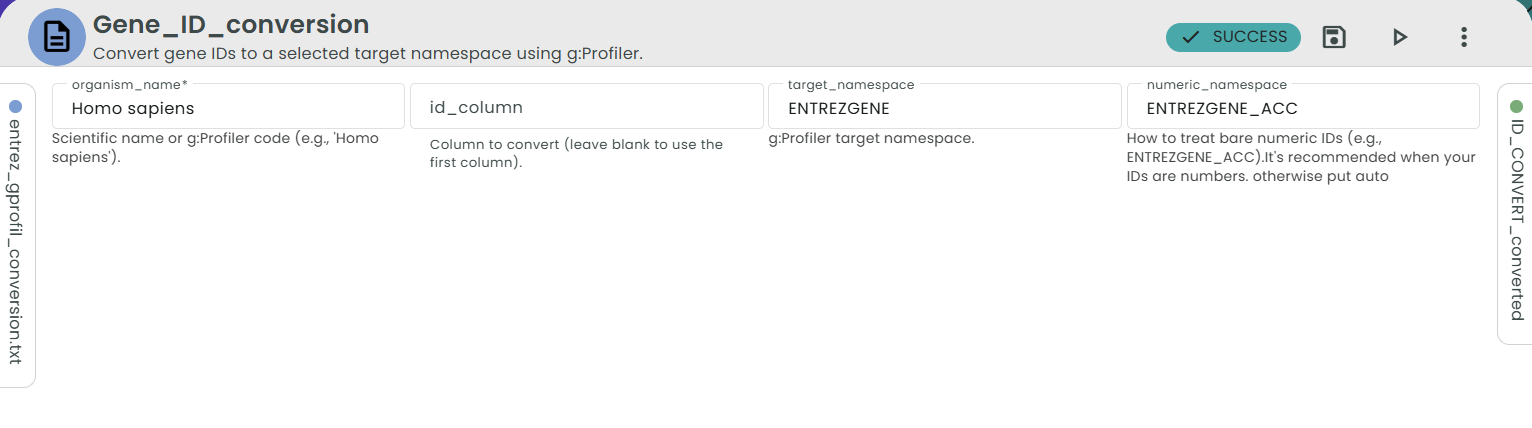
⚙️ Parameters
- organism_name Scientific name or g:Profiler code (e.g., Homo sapiens ).
- id_column (optional) Column to convert. Leave empty to use the first column in your file.
- target_namespace
Target identifier system (e.g.,
ENSG,ENTREZGENE,ENSGACC,UNIPROT, …). - numeric_namespace (optional; default auto)
How to interpret bare numeric inputs.
Choose a specific namespace (e.g.,
ENTREZGENE_ACC) when your input column is purely numbers. Use auto when your inputs are not purely numeric or already have prefixes (e.g.,ENSG…).
📤 Outputs
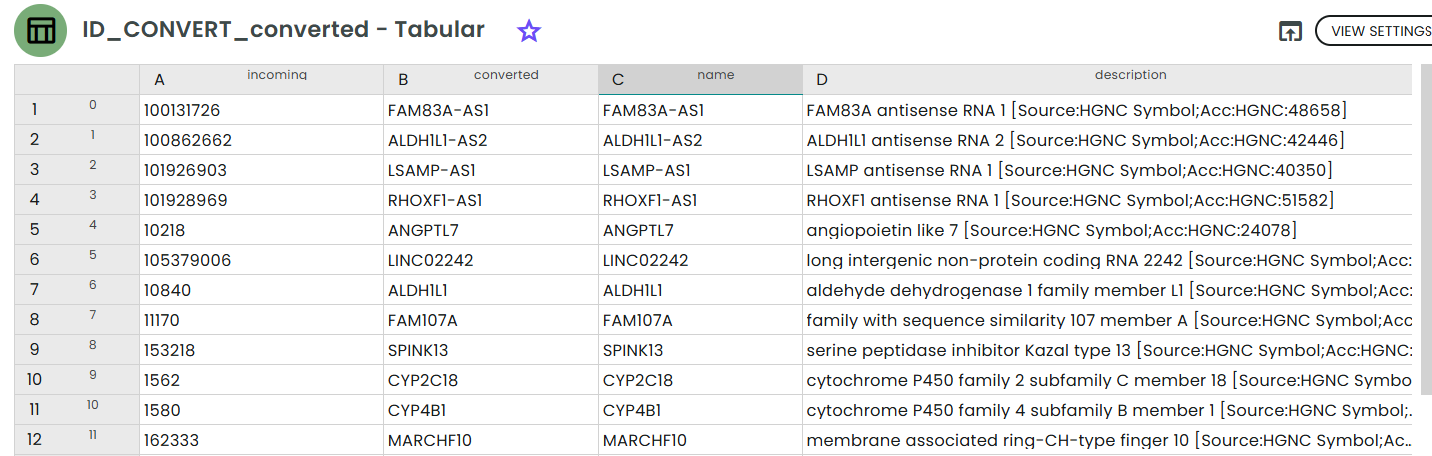
- converted_table (Table)
The conversion results; columns:
incoming– the exact input string the service receivedconverted– the mapped identifier in your target_namespacename– a convenient alias (often the preferred gene symbol)description– human-readable description if available

Comments (0)
Write a comment