Short description
GENA provides core features to create and use digital twins of cell metabolism.
How that works?
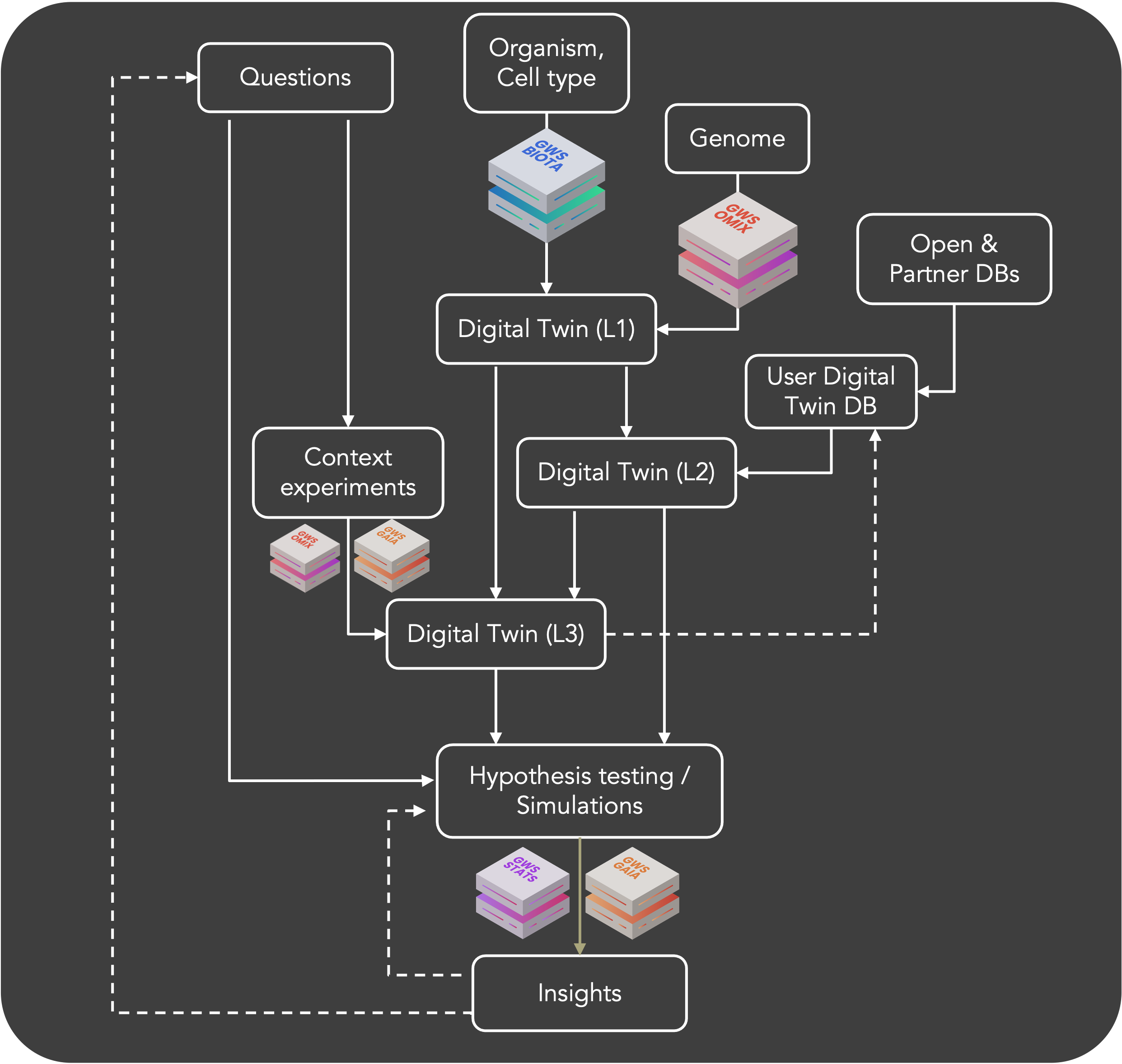
GENA goes from genomes to extract enzymatic regulations of living cells, reconstruct their whole-genome metabolic networks and create actionable digital twins. These models are simulated by computing the asymptotic distribution of metabolic fluxes into living cells, taking into account metabolic contexts. Several levels of digital twins can be created depending on the level of data used; the highest level models corresponding to models contextualized using environmental culture data (e.g. transcriptomics, proteomics, metabolomics data, etc.).
Applications and benefits
- Understand cell mechanisms of action
- Understand host-pathogen interactions
- Optimize organisms and culture media
- Optimize microbial communities
Input data
- Genome (annotated or not)
required - Context omics data (e.g. metabolomics, trascriptomics, proteomics)
optional, recommended - User Digital Twin Database
optional
Timing
- Reconstruction :
hours to up to 3 months going from a draft genome and depending on the use-case - Contextualisation :
up to 5 days (excluding data generation) - Metabolism simulation :
few seconds - Gene knockout simulation :
2-5 min
Support
- Gencovery team support (DoE, Modelling, Analysis)
- Gencovery ecosystem support (we make the connection with the right expert)
Intellectual property
- You keep IP on your proprietary data, and any related models or insights generated in Constellab™.
- You keep IP on your custom pipelines, know-how, and bricks created in Constellab™.

