Introduction
Predicting cell growth in a metabolic model holds paramount importance in understanding and manipulating cellular behavior in various fields such as biotechnology, medicine, and bioengineering. Cell growth serves as a fundamental indicator of cellular health and functionality, directly influencing the production of valuable compounds, the efficiency of metabolic processes, and even the onset and progression of diseases. By accurately predicting cell growth using metabolic models, researchers can optimize bioproduction processes, engineer microbes for industrial applications, design targeted therapies for diseases, and gain insights into the complex interplay of biochemical pathways within living organisms. Furthermore, predicting cell growth enables the identification of key metabolic pathways and regulatory mechanisms, facilitating the development of strategies to enhance productivity, resilience, and sustainability in biotechnological and biomedical applications. Thus, understanding and predicting cell growth through metabolic modeling serve as cornerstone elements in advancing our knowledge and harnessing the potential of cellular systems for a wide range of beneficial purposes.
In Constellab, you can easily predict the cell growth of your organism, as we will see in this use case.
Prerequisites
First, you need to import a metabolic network, with its context if necessary. Here we will use the e.coli model from Bigg.
Prediction of organism growth
Flux Balance Analysis (FBA)
You can predict the organism growth using the FBA Task.
Flux Balance Analysis (FBA) is a computational technique widely used in systems biology to predict the flow of metabolites through biochemical pathways within a cellular network. It operates under the assumption of steady-state conditions, where the rates of all biochemical reactions in the network are balanced. FBA employs linear programming to optimize an objective function, typically representing a cellular objective such as growth rate or biomass production, subject to constraints derived from stoichiometric relationships, enzyme capacities, and environmental conditions. By solving these optimization problems, FBA can predict metabolic flux distributions, identify metabolic bottlenecks, and suggest genetic manipulations for optimizing cellular behavior.
Once your twin is built, you can connect it to the FBA task:
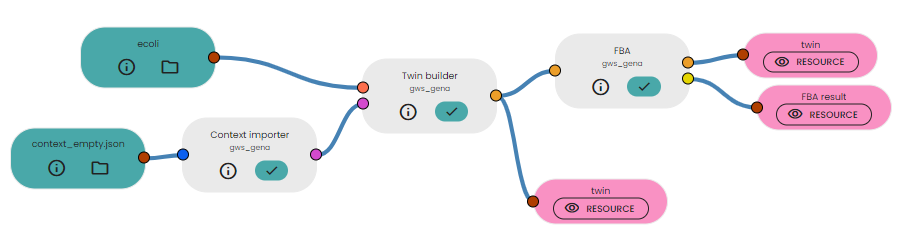
In the parameters, set the optimisation of the biomass to maximise.
In output, you will get the estimated fluxes for each reaction and above all the estimated cell growth prediction. By clicking on "twin" in the output resources, then on "summary", you can find the estimated value of biomass production:

The estimated cell growth is positive, so the organism is alive!
An alternative to the Flux balance Analysis is the Flux Variability Analysis (FVA).
Flux Variability Analysis (FVA)
Flux Variability Analysis (FVA) is a computational technique used in metabolic modeling to explore the range of flux values that can be achieved by each reaction within a metabolic network under given constraints. By systematically varying the flux of individual reactions while keeping the network in a steady state, FVA determines the maximum and minimum possible flux values for each reaction. This information is valuable for understanding the flexibility and robustness of metabolic networks, identifying critical reactions, and predicting metabolic phenotypes. FVA is commonly employed in constraint-based modeling approaches such as Flux Balance Analysis (FBA) and is a powerful tool for studying metabolic engineering, drug target identification, and metabolic pathway optimization.
Below, we applied FVA to the e.coli model, the parameters are the same as for FBA Task:
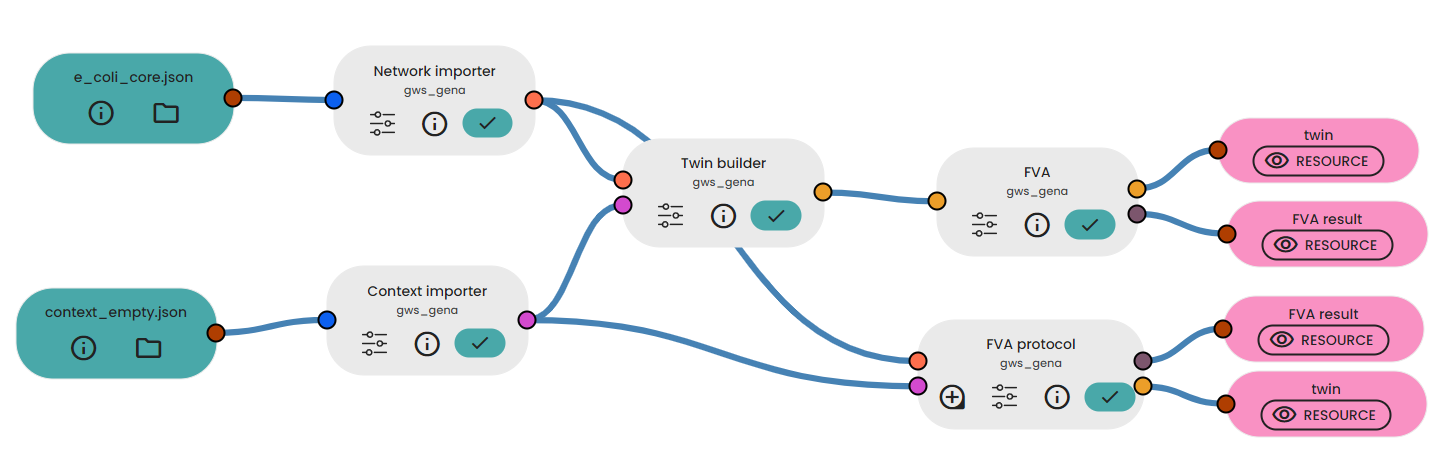
In the output we get a flux table with the minimum and maximum values for each flux. In particular, we can have an estimate of the value of biomass:


