Short description
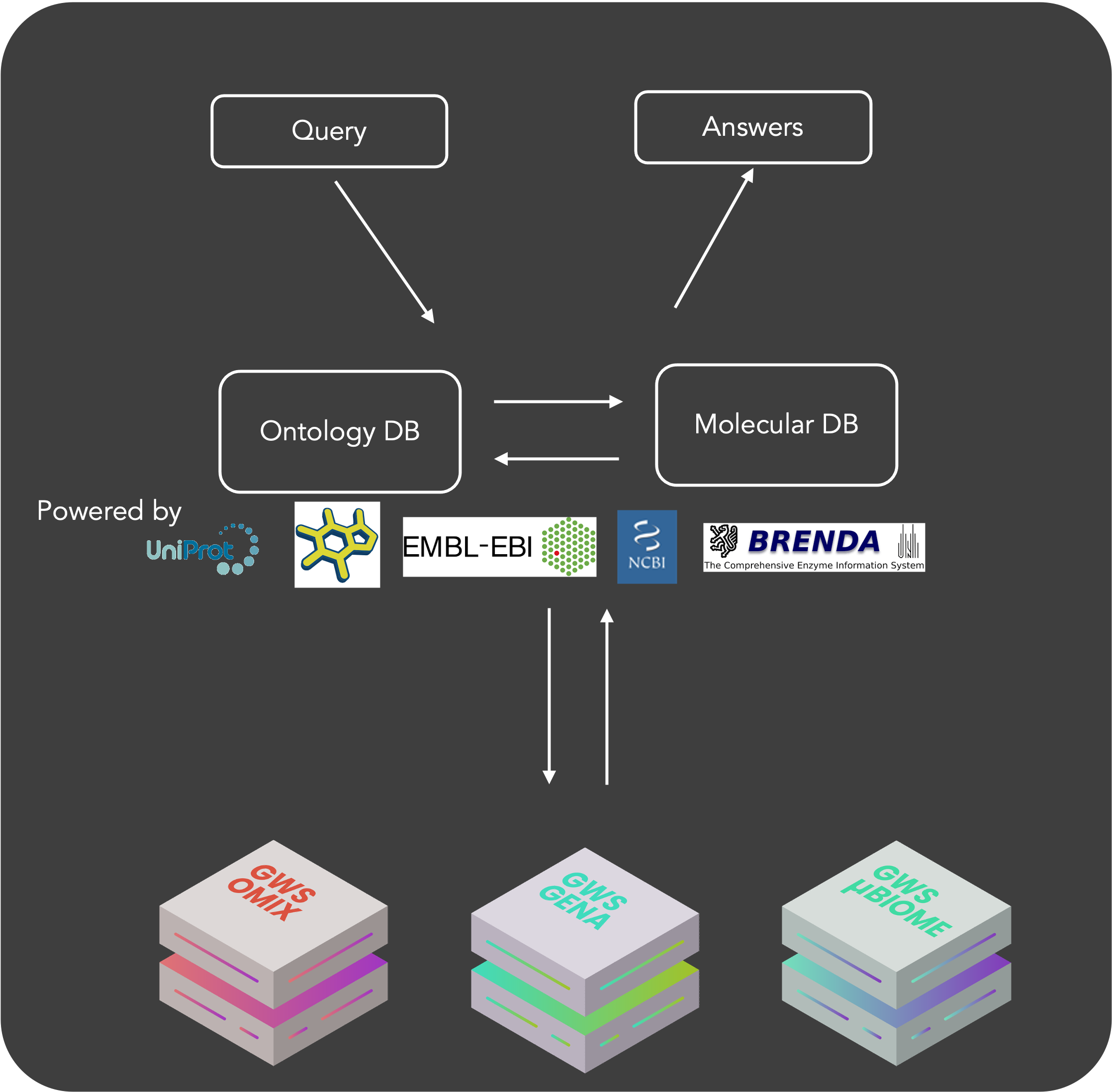
BIOTA is a unified and structured collection of omics data collected from official open European (EMBL-EBI) and NCBI taxonomy knowledge bases.
How that works?
BIOTA connect two layers of data: an ontology and molecular data manually curated by academic groups an available in European (EMBL-EBI) and NCBI taxonomy knowledge bases (taxonomy, enzymes, metabolites, pathways, …). It is dedicated to all Gencovery Web Services, particularly to the conception and use of digital twins of cell metabolism (e.g. GENA brick). So, more than 2 M organisms are referenced in BIOTA with their enzymes characteristics. BIOTA is a Gencovery open-data initiative. We are committed to keep these data open, sustainably and easily accessible to any academics and industry scientists around the World.
Applications and benefits
- Functional interpretation of bioinformatics results
- Manual querying of omics knowledge
- Automatic querying of omics knowledge for data analysis
Input data
No input data required. Everything is provided by Gencovery team.
Timing
Not applicable
Support
Gencovery team support
Intellectual Property
- BIOTA is open data and accessible to everyone (an open service dedicated to BIOTA will be available soon).
- You keep full IP on any data generated using BIOTA

